Comprehensive Assessment of Biomolecular Interactions of Morpholine-Based Mixed Ligand Cu(II) and Zn(II) Complexes of 2,2'-Bipyridine as Potential Anticancer and SARS-CoV-2 Agents: A Synergistic Experimental and Structure-Based Virtual Screening
- PMID: 36545430
- PMCID: PMC9763021
- DOI: 10.1155/2022/6987806
Comprehensive Assessment of Biomolecular Interactions of Morpholine-Based Mixed Ligand Cu(II) and Zn(II) Complexes of 2,2'-Bipyridine as Potential Anticancer and SARS-CoV-2 Agents: A Synergistic Experimental and Structure-Based Virtual Screening
Abstract
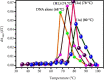
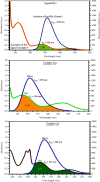
A new class of pharmacologically active mixed-ligand complexes (1a-2a) [MII(L)2 (bpy)], where L = 2-(4-morpholinobenzylideneamino)phenol), bpy = 2,2'-bipyridine, MII = Cu (1a), and Zn (2a), were assigned an octahedral geometry by analytical and spectral measurements. Gel electrophoresis showed that complex (1a) demonstrated the complete DNA cleavage mediated by H2O2. The overall DNA-binding constants observed from UV-vis, fluorometric, hydrodynamic, and electrochemical titrations were in the following sequence: (1a) > (2a) > (HL), which suggests that the complexes might intercalate DNA, a possibility that is further supported by the biothermodynamic characteristics. The binding constant results of BSA by electronic absorption and fluorometric titration demonstrate that complex (1a) exhibits the highest binding effectiveness among others, which means that all compounds could interact with BSA through a static approach, additionally supported by FRET measurements. Density FunctionalTheory (DFT) and molecular docking calculations were relied on to unveil the electronic structure, reactivity, and interacting capability of all substances with DNA, BSA, and SARS-CoV-2 main protease (Mpro). These observed binding energies fell within the following ranges: -7.7 to -8.6, -7.2 to -10.2, and -6.7 to -8.2 kcal/mol, respectively. The higher reactivity of the complexes compared to free ligand is supported by the Frontier MolecularOrbital (FMO) theory. The in vitro antibacterial, cytotoxic, and radical scavenging characteristics revealed that complex (1a) has the best biological efficacy compared to others. This is encouraged because all experimental findings are closely correlated with the theoretical measurements.
Copyright © 2022 Karunganathan Sakthikumar et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest in this work.
Figures











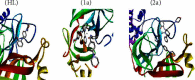



References
-
- Utthra P. P., Raman N. Probing the potency of triazole tethered Schiff base complexes and the effect of substituents on their biological attributes. International Journal of Biological Macromolecules . 2018;116:194–207. - PubMed
-
- Kumar G., Devi S., Johari R., Kumar D. Synthesis, spectral characterization and antimicrobial evaluation of Schiff base Cr (III), Mn (III) and Fe (III) macrocyclic complexes. European Journal of Medicinal Chemistry . 2012;52:269–274. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

