Recombineering in Non-Model Bacteria
- PMID: 36546891
- PMCID: PMC9793987
- DOI: 10.1002/cpz1.605
Recombineering in Non-Model Bacteria
Abstract
The technology of recombineering, in vivo genetic engineering, was initially developed in Escherichia coli and uses bacteriophage-encoded homologous recombination proteins to efficiently recombine DNA at short homologies (35 to 50 nt). Because the technology is homology driven, genomic DNA can be modified precisely and independently of restriction site location. Recombineering uses linear DNA substrates that are introduced into the cell by electroporation; these can be PCR products, synthetic double-strand DNA (dsDNA), or single-strand DNA (ssDNA). Here we describe the applications, challenges, and factors affecting ssDNA and dsDNA recombineering in a variety of non-model bacteria, both Gram-negative and -positive, and recent breakthroughs in the field. We list different microbes in which the widely used phage λ Red and Rac RecET recombination systems have been used for in vivo genetic engineering. New homologous ssDNA and dsDNA recombineering systems isolated from non-model bacteria are also described. The Basic Protocol outlines a method for ssDNA recombineering in the non-model species of Shewanella. The Alternate Protocol describes the use of CRISPR/Cas as a counter-selection system in conjunction with recombineering to enhance recovery of recombinants. We provide additional background information, pertinent considerations for experimental design, and parameters critical for success. The design of ssDNA oligonucleotides (oligos) and various internet-based tools for oligo selection from genome sequences are also described, as is the use of oligo-mediated recombination. This simple form of genome editing uses only ssDNA oligo(s) and does not require an exogenous recombination system. The information presented here should help researchers identify a recombineering system suitable for their microbe(s) of interest. If no system has been characterized for a specific microbe, researchers can find guidance in developing a recombineering system from scratch. We provide a flowchart of decision-making paths for strategically applying annealase-dependent or oligo-mediated recombination in non-model and undomesticated bacteria. © 2022 Wiley Periodicals LLC. This article has been contributed to by U.S. Government employees and their work is in the public domain in the USA. Basic Protocol: ssDNA recombineering in Shewanella species Alternate Protocol: ssDNA recombineering coupled to CRISPR/Cas9 in Shewanella species.
Keywords: RecET; annealase; non-model bacteria; recombineering; λ Red.
© 2022 Wiley Periodicals LLC. This article has been contributed to by U.S. Government employees and their work is in the public domain in the USA.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT:
Authors declare no conflict of interest.
Figures




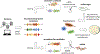
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

