RINGs, DUBs and Abnormal Brain Growth-Histone H2A Ubiquitination in Brain Development and Disease
- PMID: 36547251
- PMCID: PMC9778336
- DOI: 10.3390/epigenomes6040042
RINGs, DUBs and Abnormal Brain Growth-Histone H2A Ubiquitination in Brain Development and Disease
Abstract
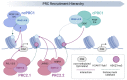
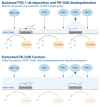
During mammalian neurodevelopment, signaling pathways converge upon transcription factors (TFs) to establish appropriate gene expression programmes leading to the production of distinct neural and glial cell types. This process is partially regulated by the dynamic modulation of chromatin states by epigenetic systems, including the polycomb group (PcG) family of co-repressors. PcG proteins form multi-subunit assemblies that sub-divide into distinct, yet functionally related families. Polycomb repressive complexes 1 and 2 (PRC1 and 2) modify the chemical properties of chromatin by covalently modifying histone tails via H2A ubiquitination (H2AK119ub1) and H3 methylation, respectively. In contrast to the PRCs, the Polycomb repressive deubiquitinase (PR-DUB) complex removes H2AK119ub1 from chromatin through the action of the C-terminal hydrolase BAP1. Genetic screening has identified several PcG mutations that are causally associated with a range of congenital neuropathologies associated with both localised and/or systemic growth abnormalities. As PRC1 and PR-DUB hold opposing functions to control H2AK119ub1 levels across the genome, it is plausible that such neurodevelopmental disorders arise through a common mechanism. In this review, we will focus on advancements regarding the composition and opposing molecular functions of mammalian PRC1 and PR-DUB, and explore how their dysfunction contributes to the emergence of neurodevelopmental disorders.
Keywords: H2AK119ub1; chromatin; chromosomal architecture; epigenetics; histone modifications; neurodevelopment; neurodevelopmental disorders; polycomb.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

