Multispecies biofilm architecture determines bacterial exposure to phages
- PMID: 36548227
- PMCID: PMC9778933
- DOI: 10.1371/journal.pbio.3001913
Multispecies biofilm architecture determines bacterial exposure to phages
Abstract
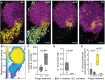
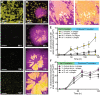
Numerous ecological interactions among microbes-for example, competition for space and resources, or interaction among phages and their bacterial hosts-are likely to occur simultaneously in multispecies biofilm communities. While biofilms formed by just a single species occur, multispecies biofilms are thought to be more typical of microbial communities in the natural environment. Previous work has shown that multispecies biofilms can increase, decrease, or have no measurable impact on phage exposure of a host bacterium living alongside another species that the phages cannot target. The reasons underlying this variability are not well understood, and how phage-host encounters change within multispecies biofilms remains mostly unexplored at the cellular spatial scale. Here, we study how the cellular scale architecture of model 2-species biofilms impacts cell-cell and cell-phage interactions controlling larger scale population and community dynamics. Our system consists of dual culture biofilms of Escherichia coli and Vibrio cholerae under exposure to T7 phages, which we study using microfluidic culture, high-resolution confocal microscopy imaging, and detailed image analysis. As shown previously, sufficiently mature biofilms of E. coli can protect themselves from phage exposure via their curli matrix. Before this stage of biofilm structural maturity, E. coli is highly susceptible to phages; however, we show that these bacteria can gain lasting protection against phage exposure if they have become embedded in the bottom layers of highly packed groups of V. cholerae in co-culture. This protection, in turn, is dependent on the cell packing architecture controlled by V. cholerae biofilm matrix secretion. In this manner, E. coli cells that are otherwise susceptible to phage-mediated killing can survive phage exposure in the absence of de novo resistance evolution. While co-culture biofilm formation with V. cholerae can confer phage protection to E. coli, it comes at the cost of competing with V. cholerae and a disruption of normal curli-mediated protection for E. coli even in dual species biofilms grown over long time scales. This work highlights the critical importance of studying multispecies biofilm architecture and its influence on the community dynamics of bacteria and phages.
Copyright: © 2022 Winans et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Krause J, Ruxton GD. Living in Groups. OUP Oxford; 2002. p 228.
-
- Seneviratne CJ, Zhang CF, Samaranayake LP. Dental plaque biofilm in oral health and disease. Chin J Dent Res Off J Sci Sect Chin Stomatol Assoc CSA. 2011;14(2):87–94. - PubMed
-
- Passow U, Ziervogel K, Asper V, Diercks A. Marine snow formation in the aftermath of the Deepwater Horizon oil spill in the Gulf of Mexico. Environ Res Lett. 2012. Jul;7(3):035301.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

