Analysis of a Dengue Virus Outbreak in Rosso, Senegal 2021
- PMID: 36548675
- PMCID: PMC9781526
- DOI: 10.3390/tropicalmed7120420
Analysis of a Dengue Virus Outbreak in Rosso, Senegal 2021
Abstract
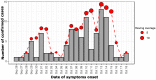
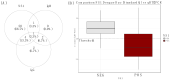
Senegal is hyperendemic for dengue. Since 2017, outbreaks have been noticed annually in many regions around the country, marked by the co-circulation of DENV1-3. On 8 October 2021, a Dengue virus outbreak in the Rosso health post (sentinel site of the syndromic surveillance network) located in the north of the country was notified to the WHO Collaborating Center for arboviruses and hemorrhagic fever viruses at Institut Pasteur de Dakar. A multidisciplinary team was then sent for epidemiological and virologic investigations. This study describes the results from investigations during an outbreak in Senegal using a rapid diagnostic test (RDT) for the combined detection of dengue virus non-structural protein 1 (NS1) and IgM/IgG. For confirmation, samples were also tested by real-time RT-PCR and IgM ELISA at the reference lab in Dakar. qRT-PCR positive samples were subjected to whole genome sequencing using nanopore technology. Virologic analysis scored 102 positives cases (RT-PCR, NS1 antigen detection and/or IgM) out of 173 enrolled patients; interestingly, virus serotyping showed that the outbreak was caused by the DENV-1, a serotype different from DENV-2 involved during the outbreak in Rosso three years earlier, indicating a serotype replacement. Nearly all field-tested NS1 positives samples were confirmed by qRT-PCR with a concordance of 92.3%. Whole genome sequencing and phylogenetic analysis of strains suggested a re-introduction in Rosso of a DENV-1 strain different to the one responsible for the outbreak in the Louga area five years before. Findings call for improved dengue virus surveillance in Senegal, with a wide deployment of DENV antigenic tests, which allow easy on-site diagnosis of suspected cases and early detection of outbreaks. This work highlights the need for continuous monitoring of circulating serotypes which is crucial for a better understanding of viral epidemiology around the country.
Keywords: DENV-1; NS1 RDTs; Rosso; outbreak response; re-introduction; serotype replacement.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- WHO/TDR, editor. Dengue: Guidelines for Diagnosis, Treatment, Prevention, and Control. TDR: World Health Organization; Geneva, Switzerland: 2009. 147p New Edtion. - PubMed
-
- WHO . Global Strategy for Dengue Prevention and Control, 2012–2020. World Health Organization; Geneva, Switzerland: 2012. [(accessed on 12 September 2020)]. Available online: http://apps.who.int/iris/bitstream/10665/75303/1/9789241504034_eng.pdf.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

