Novel insights into Hodgkin lymphoma biology by single-cell analysis
- PMID: 36548960
- PMCID: PMC10646771
- DOI: 10.1182/blood.2022017147
Novel insights into Hodgkin lymphoma biology by single-cell analysis
Abstract
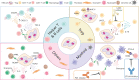
The emergence and rapid development of single-cell technologies mark a paradigm shift in cancer research. Various technology implementations represent powerful tools to understand cellular heterogeneity, identify minor cell populations that were previously hard to detect and define, and make inferences about cell-to-cell interactions at single-cell resolution. Applied to lymphoma, recent advances in single-cell RNA sequencing have broadened opportunities to delineate previously underappreciated heterogeneity of malignant cell differentiation states and presumed cell of origin, and to describe the composition and cellular subsets in the ecosystem of the tumor microenvironment (TME). Clinical deployment of an expanding armamentarium of immunotherapy options that rely on targets and immune cell interactions in the TME emphasizes the requirement for a deeper understanding of immune biology in lymphoma. In particular, classic Hodgkin lymphoma (CHL) can serve as a study paradigm because of its unique TME, featuring infrequent tumor cells among numerous nonmalignant immune cells with significant interpatient and intrapatient variability. Synergistic to advances in single-cell sequencing, multiplexed imaging techniques have added a new dimension to describing cellular cross talk in various lymphoma entities. Here, we comprehensively review recent progress using novel single-cell technologies with an emphasis on the TME biology of CHL as an application field. The described technologies, which are applicable to peripheral blood, fresh tissues, and formalin-fixed samples, hold the promise to accelerate biomarker discovery for novel immunotherapeutic approaches and to serve as future assay platforms for biomarker-informed treatment selection, including immunotherapies.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: C.S. received honoraria from Seattle Genetics, AbbVie, and Bayer, and holds research funding from Epizyme and Trillium Therapeutics Inc. T.A. declares no competing financial interests.
Figures



References
-
- Cohen M, Giladi A, Gorki AD, et al. Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell. 2018;175(4):1031–1044.e1018. - PubMed
-
- Zheng C, Zheng L, Yoo JK, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169(7):1342–1356.e1316. - PubMed
-
- Aoki T, Chong LC, Takata K, et al. Single-cell transcriptome analysis reveals disease-defining T-cell subsets in the tumor microenvironment of classic Hodgkin lymphoma. Cancer Discov. 2020;10(3):406–421. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

