Label-Free Analysis of Binding and Inhibition of SARS-Cov-19 Spike Proteins to ACE2 Receptor with ACE2-Derived Peptides by Surface Plasmon Resonance
- PMID: 36550079
- PMCID: PMC9797021
- DOI: 10.1021/acsabm.2c00832
Label-Free Analysis of Binding and Inhibition of SARS-Cov-19 Spike Proteins to ACE2 Receptor with ACE2-Derived Peptides by Surface Plasmon Resonance
Abstract
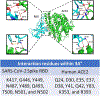
SARS-CoV-2 has been shown to enter and infect human cells via interactions between spike protein (S glycoprotein) and angiotensin-converting enzyme 2 (ACE2). As such, it may be possible to suppress the infection of the virus via the blocking of this binding interaction through the use of specific peptides that can mimic the human ACE 2 peptidase domain (PD) α 1-helix. Herein, we report the use of competitive assays along with surface plasmon resonance (SPR) to investigate the effect of peptide sequence and length on spike protein inhibition. The characterization of these binding interactions helps us understand the mechanisms behind peptide-based viral blockage and develop SPR methodologies to quickly screen disease inhibitors. This work not only helps further our understanding of the important biological interactions involved in viral inhibition but will also aid in future studies that focus on the development of therapeutics and drug options. Two peptides of different sequence lengths, [30-42] and [22-44], based on the α 1-helix of ACE2 PD were selected for this fundamental investigation. In addition to characterizing their inhibitory behavior, we also identified the critical amino acid residues of the RBD/ACE2-derived peptides by combining experimental results and molecular docking modeling. While both investigated peptides were found to effectively block the RBD residues known to bind to ACE2 PD, our investigation showed that the shorter peptide was able to reach a maximal inhibition at lower concentrations. These inhibition results matched with molecular docking models and indicated that peptide length and composition are key in the development of an effective peptide for inhibiting biophysical interactions. The work presented here emphasizes the importance of inhibition screening and modeling, as longer peptides are not always more effective.
Keywords: COVID-19; SARS-CoV-2; biosensing; molecular docking; peptide inhibitors; surface plasmon resonance.
Figures



References
-
- WHO COVID-19 Dashboard. Geneva: World Health Organization 2020: Available online: https://covid19.who.int/ Sept. 9th 2022.
-
- Mahmud N; Anik MI; Hossain MK; Khan MI; Uddin S; Ashrafuzzaman M; Rahaman MM, Advances in Nanomaterial-Based Platforms to Combat COVID-19: Diagnostics, Preventions, Therapeutics, and Vaccine Developments. ACS Applied Bio Materials 2022. 5, 6, 2431–2460. - PubMed
-
- Karmacharya M; Kumar S; Gulenko O; Cho Y-K, Advances in facemasks during the COVID-19 pandemic era. ACS Applied Bio Materials 2021, 4 (5), 3891–3908. - PubMed
-
- Yadav S; Sadique MA; Ranjan P; Kumar N; Singhal A; Srivastava AK; Khan R, SERS Based Lateral Flow Immunoassay for Point-of-Care Detection of SARS-CoV-2 in Clinical Samples. ACS Appl Bio Mater 2021, 4 (4), 2974–2995. - PubMed
-
- Pramanik A; Mayer J; Sinha SS; Sharma PC; Patibandla S; Gao Y; Corby LR; Bates JT; Bierdeman MA; Tandon R; Seshadri R; Ray PC, Human ACE2 Peptide-Attached Plasmonic-Magnetic Heterostructure for Magnetic Separation, Surface Enhanced Raman Spectroscopy Identification, and Inhibition of Different Variants of SARS-CoV-2 Infections. ACS Appl Bio Mater 2022. 5, 9, 4454–4464. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
