Transcriptome analysis reveals differential transcription in tomato (Solanum lycopersicum) following inoculation with Ralstonia solanacearum
- PMID: 36550145
- PMCID: PMC9780229
- DOI: 10.1038/s41598-022-26693-y
Transcriptome analysis reveals differential transcription in tomato (Solanum lycopersicum) following inoculation with Ralstonia solanacearum
Abstract
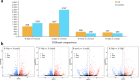
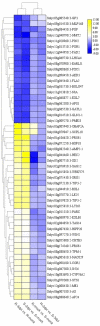
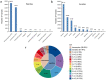
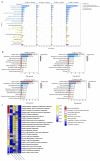
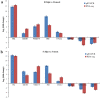
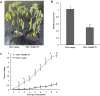
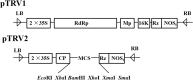
Tomato (Solanum lycopersicum L.) is a major Solanaceae crop worldwide and is vulnerable to bacterial wilt (BW) caused by Ralstonia solanacearum during the production process. BW has become a growing concern that could enormously deplete the tomato yield from 50 to 100% and decrease the quality. Research on the molecular mechanism of tomato regulating BW resistance is still limited. In this study, two tomato inbred lines (Hm 2-2, resistant to BW; and BY 1-2, susceptible to BW) were used to explore the molecular mechanism of tomato in response to R. solanacearum infection by RNA-sequencing (RNA-seq) technology. We identified 1923 differentially expressed genes (DEGs) between Hm 2-2 and BY 1-2 after R. solanacearum inoculation. Among these DEGs, 828 were up-regulated while 1095 were down-regulated in R-3dpi (Hm 2-2 at 3 days post-inoculation with R. solanacearum) vs. R-mock (mock-inoculated Hm 2-2); 1087 and 2187 were up- and down-regulated, respectively, in S-3dpi (BY 1-2 at 3 days post-inoculation with R. solanacearum) vs. S-mock (mock-inoculated BY 1-2). Moreover, Gene Ontology (GO) enrichment analysis revealed that the largest amount of DEGs were annotated with the Biological Process terms, followed by Cellular Component and Molecular Function terms. A total of 114, 124, 85, and 89 regulated (or altered) pathways were identified in R-3dpi vs. R-mock, S-3dpi vs. S-mock, R-mock vs. S-mock, and R-3dpi vs. S-3dpi comparisons, respectively, by Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway analysis. These clarified the molecular function and resistance pathways of DEGs. Furthermore, quantitative RT-PCR (qRT-PCR) analysis confirmed the expression patterns of eight randomly selected DEGs, which suggested that the RNA-seq results were reliable. Subsequently, in order to further verify the reliability of the transcriptome data and the accuracy of qRT-PCR results, WRKY75, one of the eight DEGs was silenced by virus-induced gene silencing (VIGS) and the defense response of plants to R. solanacearum infection was analyzed. In conclusion, the findings of this study provide profound insight into the potential mechanism of tomato in response to R. solanacearum infection, which lays an important foundation for future studies on BW.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

