Integrative genomic analysis of childhood acute lymphoblastic leukaemia lacking a genetic biomarker in the UKALL2003 clinical trial
- PMID: 36550215
- PMCID: PMC9991913
- DOI: 10.1038/s41375-022-01799-4
Integrative genomic analysis of childhood acute lymphoblastic leukaemia lacking a genetic biomarker in the UKALL2003 clinical trial
Abstract
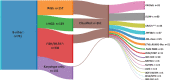
Incorporating genetics into risk-stratification for treatment of childhood B-progenitor acute lymphoblastic leukaemia (B-ALL) has contributed significantly to improved survival. In about 30% B-ALL (B-other-ALL) without well-established chromosomal changes, new genetic subtypes have recently emerged, yet their true prognostic relevance largely remains unclear. We integrated next generation sequencing (NGS): whole genome sequencing (WGS) (n = 157) and bespoke targeted NGS (t-NGS) (n = 175) (overlap n = 36), with existing genetic annotation in a representative cohort of 351 B-other-ALL patients from the childhood ALL trail, UKALL2003. PAX5alt was most frequently observed (n = 91), whereas PAX5 P80R mutations (n = 11) defined a distinct PAX5 subtype. DUX4-r subtype (n = 80) was defined by DUX4 rearrangements and/or ERG deletions. These patients had a low relapse rate and excellent survival. ETV6::RUNX1-like subtype (n = 21) was characterised by multiple abnormalities of ETV6 and IKZF1, with no reported relapses or deaths, indicating their excellent prognosis in this trial. An inferior outcome for patients with ABL-class fusions (n = 25) was confirmed. Integration of NGS into genomic profiling of B-other-ALL within a single childhood ALL trial, UKALL2003, has shown the added clinical value of NGS-based approaches, through improved accuracy in detection and classification into the range of risk stratifying genetic subtypes, while validating their prognostic significance.
© 2022. The Author(s).
Conflict of interest statement
MTR, DRB, JFP, and ZK are employees of Illumina, a public company that develops and markets systems for genetic analysis. The remaining authors declare no competing interests.
Figures



References
-
- Moorman AV, Ensor HM, Richards SM, Chilton L, Schwab C, Kinsey SE, et al. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: Results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010;11:429–38. doi: 10.1016/S1470-2045(10)70066-8. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

