Virus-assisted directed evolution of enhanced suppressor tRNAs in mammalian cells
- PMID: 36550276
- PMCID: PMC9855281
- DOI: 10.1038/s41592-022-01706-w
Virus-assisted directed evolution of enhanced suppressor tRNAs in mammalian cells
Abstract
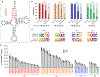
Site-specific incorporation of unnatural amino acids (Uaas) in living cells relies on engineered aminoacyl-transfer RNA synthetase-tRNA pairs borrowed from a distant domain of life. Such heterologous suppressor tRNAs often have poor intrinsic activity, presumably due to suboptimal interaction with a non-native translation system. This limitation can be addressed in Escherichia coli using directed evolution. However, no suitable selection system is currently available to do the same in mammalian cells. Here we report virus-assisted directed evolution of tRNAs (VADER) in mammalian cells, which uses a double-sieve selection scheme to facilitate single-step enrichment of active yet orthogonal tRNA mutants from naive libraries. Using VADER we developed improved mutants of Methanosarcina mazei pyrrolysyl-tRNA, as well as a bacterial tyrosyl-tRNA. We also show that the higher activity of the most efficient mutant pyrrolysyl-tRNA is specific for mammalian cells, alluding to an improved interaction with the unique mammalian translation apparatus.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests statement
A patent application has been submitted on the improved tRNA mutants on which A.C, D.J., and R.E.K. are coinventors. A.C. is a senior advisor at BrickBio, Inc. R.L.H., Z.Z., B.S., X.C., K.M., Z.H., M.P., J.A., and T.v.O. have no competing interests.
Figures
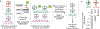



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

