Engineering Human Mesenchymal Bodies in a Novel 3D-Printed Microchannel Bioreactor for Extracellular Vesicle Biogenesis
- PMID: 36551001
- PMCID: PMC9774207
- DOI: 10.3390/bioengineering9120795
Engineering Human Mesenchymal Bodies in a Novel 3D-Printed Microchannel Bioreactor for Extracellular Vesicle Biogenesis
Abstract
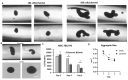
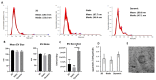
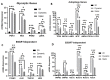
Human Mesenchymal Stem Cells (hMSCs) and their derived products hold potential in tissue engineering and as therapeutics in a wide range of diseases. hMSCs possess the ability to aggregate into "spheroids", which has been used as a preconditioning technique to enhance their therapeutic potential by upregulating stemness, immunomodulatory capacity, and anti-inflammatory and pro-angiogenic secretome. Few studies have investigated the impact on hMSC aggregate properties stemming from dynamic and static aggregation techniques. hMSCs' main mechanistic mode of action occur through their secretome, including extracellular vesicles (EVs)/exosomes, which contain therapeutically relevant proteins and nucleic acids. In this study, a 3D printed microchannel bioreactor was developed to dynamically form hMSC spheroids and promote hMSC condensation. In particular, the manner in which dynamic microenvironment conditions alter hMSC properties and EV biogenesis in relation to static cultures was assessed. Dynamic aggregation was found to promote autophagy activity, alter metabolism toward glycolysis, and promote exosome/EV production. This study advances our knowledge on a commonly used preconditioning technique that could be beneficial in wound healing, tissue regeneration, and autoimmune disorders.
Keywords: 3-D aggregates; extracellular vesicle biogenesis; human mesenchymal stem cells; microchannel; wave motion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Samadian S., Karbalaei A., Pourmadadi M., Yazdian F., Rashedi H., Omidi M., Malmir S. A novel alginate-gelatin microcapsule to enhance bone differentiation of mesenchymal stem cells. Int. J. Polym. Mater. 2021;71:395–402. doi: 10.1080/00914037.2020.1848828. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

