Correlative Raman-Electron-Light (CREL) Microscopy Analysis of Lipid Droplets in Melanoma Cancer Stem Cells
- PMID: 36551069
- PMCID: PMC9776032
- DOI: 10.3390/bios12121102
Correlative Raman-Electron-Light (CREL) Microscopy Analysis of Lipid Droplets in Melanoma Cancer Stem Cells
Abstract
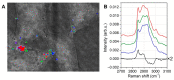
Among all neoplasms, melanoma is characterized by a very high percentage of cancer stem cells (CSCs). Several markers have been proposed for their identification, and lipid droplets (LDs) are among them. Different techniques are used for their characterization such as mass spectrometry, imaging techniques, and vibrational spectroscopies. Some emerging experimental approaches for the study of LDs are represented by correlative light-electron microscopy and by correlative Raman imaging-scanning electron microscopy (SEM). Based on these scientific approaches, we developed a novel methodology (CREL) by combining Raman micro-spectroscopy, confocal fluorescence microscopy, and SEM coupled with an energy-dispersive X-ray spectroscopy module. This procedure correlated cellular morphology, chemical properties, and spatial distribution from the same region of interest, and in this work, we presented the application of CREL for the analysis of LDs within patient-derived melanoma CSCs (MCSCs).
Keywords: Raman micro-imaging; correlative microscopy; electron microscopy; lipid droplets; melanoma cancer stem cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

