Hub Genes in Non-Small Cell Lung Cancer Regulatory Networks
- PMID: 36551208
- PMCID: PMC9776006
- DOI: 10.3390/biom12121782
Hub Genes in Non-Small Cell Lung Cancer Regulatory Networks
Abstract
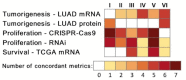
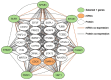
There are currently no accurate biomarkers for optimal treatment selection in early-stage non-small cell lung cancer (NSCLC). Novel therapeutic targets are needed to improve NSCLC survival outcomes. This study systematically evaluated the association between genome-scale regulatory network centralities and NSCLC tumorigenesis, proliferation, and survival in early-stage NSCLC patients. Boolean implication networks were used to construct multimodal networks using patient DNA copy number variation, mRNA, and protein expression profiles. T statistics of differential gene/protein expression in tumors versus non-cancerous adjacent tissues, dependency scores in in vitro CRISPR-Cas9/RNA interference (RNAi) screening of human NSCLC cell lines, and hazard ratios in univariate Cox modeling of the Cancer Genome Atlas (TCGA) NSCLC patients were correlated with graph theory centrality metrics. Hub genes in multi-omics networks involving gene/protein expression were associated with oncogenic, proliferative potentials and poor patient survival outcomes (p < 0.05, Pearson's correlation). Immunotherapy targets PD1, PDL1, CTLA4, and CD27 were ranked as top hub genes within the 10th percentile in most constructed multi-omics networks. BUB3, DNM1L, EIF2S1, KPNB1, NMT1, PGAM1, and STRAP were discovered as important hub genes in NSCLC proliferation with oncogenic potential. These results support the importance of hub genes in NSCLC tumorigenesis, proliferation, and prognosis, with implications in prioritizing therapeutic targets to improve patient survival outcomes.
Keywords: CRISPR-Cas9; RNAi; biomarkers; hub genes; multi-omics networks; non-small cell lung cancer; patient survival; proliferation; therapeutic targets.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Inferencing Bulk Tumor and Single-Cell Multi-Omics Regulatory Networks for Discovery of Biomarkers and Therapeutic Targets.Cells. 2022 Dec 26;12(1):101. doi: 10.3390/cells12010101. Cells. 2022. PMID: 36611894 Free PMC article. Review.
-
Immune-Omics Networks of CD27, PD1, and PDL1 in Non-Small Cell Lung Cancer.Cancers (Basel). 2021 Aug 26;13(17):4296. doi: 10.3390/cancers13174296. Cancers (Basel). 2021. PMID: 34503105 Free PMC article.
-
A Multi-Omics Network of a Seven-Gene Prognostic Signature for Non-Small Cell Lung Cancer.Int J Mol Sci. 2021 Dec 25;23(1):219. doi: 10.3390/ijms23010219. Int J Mol Sci. 2021. PMID: 35008645 Free PMC article.
-
Multi-Omics Immune Interaction Networks in Lung Cancer Tumorigenesis, Proliferation, and Survival.Int J Mol Sci. 2022 Nov 29;23(23):14978. doi: 10.3390/ijms232314978. Int J Mol Sci. 2022. PMID: 36499305 Free PMC article.
-
Using biological information to analyze potential miRNA-mRNA regulatory networks in the plasma of patients with non-small cell lung cancer.BMC Cancer. 2022 Mar 21;22(1):299. doi: 10.1186/s12885-022-09281-1. BMC Cancer. 2022. PMID: 35313857 Free PMC article.
Cited by
-
Identification of a central network hub of key prognostic genes based on correlation between transcriptomics and survival in patients with metastatic solid tumors.Ther Adv Med Oncol. 2024 Oct 17;16:17588359241289200. doi: 10.1177/17588359241289200. eCollection 2024. Ther Adv Med Oncol. 2024. PMID: 39429467 Free PMC article.
-
Inferencing Bulk Tumor and Single-Cell Multi-Omics Regulatory Networks for Discovery of Biomarkers and Therapeutic Targets.Cells. 2022 Dec 26;12(1):101. doi: 10.3390/cells12010101. Cells. 2022. PMID: 36611894 Free PMC article. Review.
-
Investigation of Diagnostic and Prognostic Value of CLEC4M of Non-Small Cell Lung Carcinoma Associated with Immune Microenvironment.Int J Gen Med. 2023 Apr 15;16:1317-1332. doi: 10.2147/IJGM.S397695. eCollection 2023. Int J Gen Med. 2023. PMID: 37089135 Free PMC article.
References
-
- Lung Cancer—Non-Small Cell: Statistics. [(accessed on 23 May 2022)]. Available online: https://www.cancer.net/cancer-types/lung-cancer-non-small-cell/statistics.
-
- Hellmann M.D., Rizvi N.A., Goldman J.W., Gettinger S.N., Borghaei H., Brahmer J.R., Ready N.E., Gerber D.E., Chow L.Q., Juergens R.A., et al. Nivolumab plus ipilimumab as first-line treatment for advanced non-small-cell lung cancer (CheckMate 012): Results of an open-label, phase 1, multicohort study. Lancet Oncol. 2017;18:31–41. doi: 10.1016/S1470-2045(16)30624-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

