Degradation Mechanism of AAA+ Proteases and Regulation of Streptomyces Metabolism
- PMID: 36551276
- PMCID: PMC9775585
- DOI: 10.3390/biom12121848
Degradation Mechanism of AAA+ Proteases and Regulation of Streptomyces Metabolism
Abstract
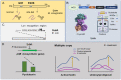
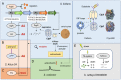
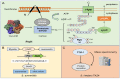
Hundreds of proteins work together in microorganisms to coordinate and control normal activity in cells. Their degradation is not only the last step in the cell's lifespan but also the starting point for its recycling. In recent years, protein degradation has been extensively studied in both eukaryotic and prokaryotic organisms. Understanding the degradation process is essential for revealing the complex regulatory network in microorganisms, as well as further artificial reconstructions and applications. This review will discuss several studies on protein quality-control family members Lon, FtsH, ClpP, the proteasome in Streptomyces, and a few classical model organisms, mainly focusing on their structure, recognition mechanisms, and metabolic influences.
Keywords: ClpP; FtsH; Lon; Streptomyces; proteasome; protein degradation; regulatory network.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Antibiotic Acyldepsipeptides Stimulate the Streptomyces Clp-ATPase/ClpP Complex for Accelerated Proteolysis.mBio. 2022 Dec 20;13(6):e0141322. doi: 10.1128/mbio.01413-22. Epub 2022 Oct 26. mBio. 2022. PMID: 36286522 Free PMC article.
-
Regulated Proteolysis in Bacteria.Annu Rev Biochem. 2018 Jun 20;87:677-696. doi: 10.1146/annurev-biochem-062917-012848. Epub 2018 Apr 12. Annu Rev Biochem. 2018. PMID: 29648875 Free PMC article. Review.
-
AAA+ ATPases in Protein Degradation: Structures, Functions and Mechanisms.Biomolecules. 2020 Apr 18;10(4):629. doi: 10.3390/biom10040629. Biomolecules. 2020. PMID: 32325699 Free PMC article. Review.
-
Proteasomes: unfoldase-assisted protein degradation machines.Biol Chem. 2019 Dec 18;401(1):183-199. doi: 10.1515/hsz-2019-0344. Biol Chem. 2019. PMID: 31665105 Review.
-
Proteolysis mediated by the membrane-integrated ATP-dependent protease FtsH has a unique nonlinear dependence on ATP hydrolysis rates.Protein Sci. 2019 Jul;28(7):1262-1275. doi: 10.1002/pro.3629. Epub 2019 May 8. Protein Sci. 2019. PMID: 31008538 Free PMC article.
Cited by
-
LexA, an SOS response repressor, activates TGase synthesis in Streptomyces mobaraensis.Front Microbiol. 2024 May 24;15:1397314. doi: 10.3389/fmicb.2024.1397314. eCollection 2024. Front Microbiol. 2024. PMID: 38855760 Free PMC article.
-
Role of Carbon, Nitrogen, Phosphate and Sulfur Metabolism in Secondary Metabolism Precursor Supply in Streptomyces spp.Microorganisms. 2024 Jul 31;12(8):1571. doi: 10.3390/microorganisms12081571. Microorganisms. 2024. PMID: 39203413 Free PMC article. Review.
-
Inhibitory effects of nafcillin and diosmin on biofilm formation by Salmonella Typhimurium.BMC Microbiol. 2024 Dec 18;24(1):522. doi: 10.1186/s12866-024-03646-1. BMC Microbiol. 2024. PMID: 39695365 Free PMC article.
-
Separation of Lysozyme-Ovotransferrin Complexes and the Cooperative Role of Their Components in Egg White.Appl Biochem Biotechnol. 2025 Jan;197(1):370-383. doi: 10.1007/s12010-024-05037-8. Epub 2024 Aug 9. Appl Biochem Biotechnol. 2025. PMID: 39120837
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

