Detection of QTLs Regulating Six Agronomic Traits of Rice Based on Chromosome Segment Substitution Lines of Common Wild Rice (Oryza rufipogon Griff.) and Mapping of qPH1.1 and qLMC6.1
- PMID: 36551278
- PMCID: PMC9775987
- DOI: 10.3390/biom12121850
Detection of QTLs Regulating Six Agronomic Traits of Rice Based on Chromosome Segment Substitution Lines of Common Wild Rice (Oryza rufipogon Griff.) and Mapping of qPH1.1 and qLMC6.1
Abstract
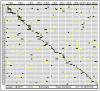
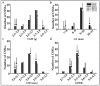
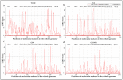
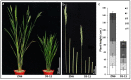
Wild rice is a primary source of genes that can be utilized to generate rice cultivars with advantageous traits. Chromosome segment substitution lines (CSSLs) are consisting of a set of consecutive and overlapping donor chromosome segments in a recipient's genetic background. CSSLs are an ideal genetic population for mapping quantitative traits loci (QTLs). In this study, 59 CSSLs from the common wild rice (Oryza rufipogon Griff.) accession DP15 under the indica rice cultivar (O. sativa L. ssp. indica) variety 93-11 background were constructed through multiple backcrosses and marker-assisted selection (MAS). Through high-throughput whole genome re-sequencing (WGRS) of parental lines, 12,565 mapped InDels were identified and designed for polymorphic molecular markers. The 59 CSSLs library covered 91.72% of the genome of common wild rice accession DP15. The DP15-CSSLs displayed variation in six economic traits including grain length (GL), grain width (GW), thousand-grain weight (TGW), grain length-width ratio (GLWR), plant height (PH), and leaf margin color (LMC), which were finally attributed to 22 QTLs. A homozygous CSSL line and a purple leave margin CSSL line were selected to construct two secondary genetic populations for the QTLs mapping. Thus, the PH-controlling QTL qPH1.1 was mapped to a region of 4.31-Mb on chromosome 1, and the LMC-controlling QTL qLMC6.1 was mapped to a region of 370-kb on chromosome 6. Taken together, these identified novel QTLs/genes from common wild rice can potentially promote theoretical knowledge and genetic applications to rice breeders worldwide.
Keywords: QTL mapping; agronomic traits; chromosome segment substitution lines (CSSLs); common wild rice (Oryza rufipogon); molecular markers; qLMC6.1; qPH1.1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Development of Chromosome Segment Substitution Lines (CSSLs) Derived from Guangxi Wild Rice (Oryza rufipogon Griff.) under Rice (Oryza sativa L.) Background and the Identification of QTLs for Plant Architecture, Agronomic Traits and Cold Tolerance.Genes (Basel). 2020 Aug 22;11(9):980. doi: 10.3390/genes11090980. Genes (Basel). 2020. PMID: 32842674 Free PMC article.
-
Construction of chromosome segment substitution lines of Dongxiang common wild rice (Oryza rufipogon Griff.) in the background of the japonica rice cultivar Nipponbare (Oryza sativa L.).Plant Physiol Biochem. 2019 Nov;144:274-282. doi: 10.1016/j.plaphy.2019.09.041. Epub 2019 Sep 25. Plant Physiol Biochem. 2019. PMID: 31593900
-
Development and characterization of chromosome segment substitution lines derived from Oryza rufipogon in the genetic background of O. sativa spp. indica cultivar 9311.BMC Genomics. 2016 Aug 9;17:580. doi: 10.1186/s12864-016-2987-5. BMC Genomics. 2016. PMID: 27507407 Free PMC article.
-
Development and use of chromosome segment substitution lines as a genetic resource for crop improvement.Theor Appl Genet. 2019 Jan;132(1):1-25. doi: 10.1007/s00122-018-3219-y. Epub 2018 Nov 27. Theor Appl Genet. 2019. PMID: 30483819 Review.
-
2Gs and plant architecture: breaking grain yield ceiling through breeding approaches for next wave of revolution in rice (Oryza sativa L.).Crit Rev Biotechnol. 2024 Feb;44(1):139-162. doi: 10.1080/07388551.2022.2112648. Epub 2022 Sep 29. Crit Rev Biotechnol. 2024. PMID: 36176065 Review.
References
-
- Descalsota-Empleo G.I., Noraziyah A.A.S., Navea I.P., Chung C., Dwiyanti M.S., Labios R.J.D., Ikmal A.M., Juanillas V.M., Inabangan-Asilo M.A., Amparado A., et al. Genetic Dissection of Grain Nutritional Traits and Leaf Blight Resistance in Rice. Genes. 2019;10:30. doi: 10.3390/genes10010030. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

