Identification and Validation of Cuproptosis-Related LncRNA Signatures in the Prognosis and Immunotherapy of Clear Cell Renal Cell Carcinoma Using Machine Learning
- PMID: 36551318
- PMCID: PMC9776244
- DOI: 10.3390/biom12121890
Identification and Validation of Cuproptosis-Related LncRNA Signatures in the Prognosis and Immunotherapy of Clear Cell Renal Cell Carcinoma Using Machine Learning
Abstract
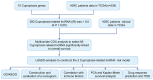
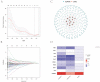
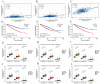
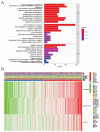
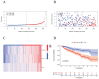
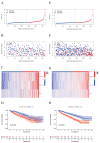
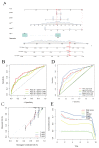
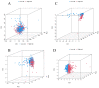
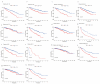
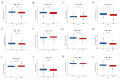
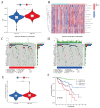
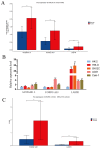
(1) Objective: We aimed to mine cuproptosis-related LncRNAs with prognostic value and construct a corresponding prognostic model using machine learning. External validation of the model was performed in the ICGC database and in multiple renal cancer cell lines via qPCR. (2) Methods: TCGA and ICGC cohorts related to renal clear cell carcinoma were included. GO and KEGG analyses were conducted to determine the biological significance of differentially expressed cuproptosis-related LncRNAs (CRLRs). Machine learning (LASSO), Kaplan-Meier, and Cox analyses were conducted to determine the prognostic genes. The tumor microenvironment and tumor mutation load were further studied. TIDE and IC50 were used to evaluate the response to immunotherapy, a risk model of LncRNAs related to the cuproptosis genes was established, and the ability of this model was verified in an external independent ICGC cohort. LncRNAs were identified in normal HK-2 cells and verified in four renal cell lines via qPCR. (3) Results: We obtained 280 CRLRs and identified 66 LncRNAs included in the TCGA-KIRC cohort. Then, three hub LncRNAs (AC026401.3, FOXD2-AS1, and LASTR), which were over-expressed in the four ccRCC cell lines compared with the human renal cortex proximal tubule epithelial cell line HK-2, were identified. In the ICGC database, the expression of FOXD2-AS1 and LASTR was consistent with the qPCR and TCGA-KIRC. The results also indicated that patients with low-risk ccRCC-stratified by tumor-node metastasis stage, sex, and tumor grade-had significantly better overall survival than those with high-risk ccRCC. The predictive algorithm showed that, according to the three CRLR models, the low-risk group was more sensitive to nine target drugs (A.443654, A.770041, ABT.888, AG.014699, AMG.706, ATRA, AP.24534, axitinib, and AZ628), based on the estimated half-maximal inhibitory concentrations. In contrast, the high-risk group was more sensitive to ABT.263 and AKT inhibitors VIII and AS601245. Using the CRLR models, the correlation between the tumor immune microenvironment and cancer immunotherapy response revealed that high-risk patients are more likely to respond to immunotherapy than low-risk patients. In terms of immune marker levels, there were significant differences between the high- and low-risk groups. A high TMB score in the high-risk CRLR group was associated with worse survival, which could be a prognostic factor for KIRC. (4) Conclusions: This study elucidates the core cuproptosis-related LncRNAs, FOXD2-AS1, AC026401.3, and LASTR, in terms of potential predictive value, immunotherapeutic strategy, and outcome of ccRCC.
Keywords: TMB score; ccRCC; cuproptosis; immunotherapy; lncRNA.
Conflict of interest statement
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- HZ-2021- No. 218/Technological Project of Zunyi Science and Technology Bureau
- HZ-2021-No. 298/Technological Project of Zunyi Science and Technology Bureau
- QianKeHeZhiCheng [2022]YiBan182/Natural Science and Technology Foundation of Guizhou Province
- (2020-024)/The Special Project of Innovation and Exploration in Zunyi Medical University
- gzwkj 2021-138/The Project of Guizhou Provincial Health Commission
- gzwkj2021-517/The Project of Guizhou Provincial Health Commission
- QianJiaoHe KY Zi[2022]280 Hao/The Young Scientific and Technological Talents Growth Project of Guizhou Provincial Department of Education
- 82260106/National Natural Science Foundation of China
- gzwkj2021-539/The Project of Guizhou Provincial Health Commission
- QKHRCPT [2019] No.1332/Natural Science and Technology Foundation of Guizhou Province
LinkOut - more resources
Full Text Sources
Medical

