Discovery of Highly Trimethoprim-Resistant DfrB Dihydrofolate Reductases in Diverse Environmental Settings Suggests an Evolutionary Advantage Unrelated to Antibiotic Resistance
- PMID: 36551425
- PMCID: PMC9774602
- DOI: 10.3390/antibiotics11121768
Discovery of Highly Trimethoprim-Resistant DfrB Dihydrofolate Reductases in Diverse Environmental Settings Suggests an Evolutionary Advantage Unrelated to Antibiotic Resistance
Abstract
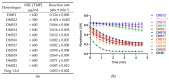
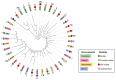
Type B dihydrofolate reductases (DfrB) are intrinsically highly resistant to the widely used antibiotic trimethoprim, posing a threat to global public health. The ten known DfrB family members have been strongly associated with genetic material related to the application of antibiotics. Several dfrB genes were associated with multidrug resistance contexts and mobile genetic elements, integrated both in chromosomes and plasmids. However, little is known regarding their presence in other environments. Here, we investigated the presence of dfrB beyond the traditional areas of enquiry by conducting metagenomic database searches from environmental settings where antibiotics are not prevalent. Thirty putative DfrB homologues that share 62 to 95% identity with characterized DfrB were identified. Expression of ten representative homologues verified trimethoprim resistance in all and dihydrofolate reductase activity in most. Contrary to samples associated with the use of antibiotics, the newly identified dfrB were rarely associated with mobile genetic elements or antibiotic resistance genes. Instead, association with metabolic enzymes was observed, suggesting an evolutionary advantage unrelated to antibiotic resistance. Our results are consistent with the hypothesis that multiple dfrB exist in diverse environments from which dfrB were mobilized into the clinically relevant resistome. Our observations reinforce the need to closely monitor their progression.
Keywords: antibiotic resistance; metagenomic database search; mobile genetic elements; multidrug resistance; type B dihydrofolate reductase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Trimethoprim resistance in surface and wastewater is mediated by contrasting variants of the dfrB gene.ISME J. 2023 Sep;17(9):1455-1466. doi: 10.1038/s41396-023-01460-7. Epub 2023 Jun 27. ISME J. 2023. PMID: 37369703 Free PMC article.
-
The Bacterial Genomic Context of Highly Trimethoprim-Resistant DfrB Dihydrofolate Reductases Highlights an Emerging Threat to Public Health.Antibiotics (Basel). 2021 Apr 13;10(4):433. doi: 10.3390/antibiotics10040433. Antibiotics (Basel). 2021. PMID: 33924456 Free PMC article.
-
A conserved SH3-like fold in diverse putative proteins tetramerizes into an oxidoreductase providing an antimicrobial resistance phenotype.Philos Trans R Soc Lond B Biol Sci. 2023 Feb 27;378(1871):20220040. doi: 10.1098/rstb.2022.0040. Epub 2023 Jan 11. Philos Trans R Soc Lond B Biol Sci. 2023. PMID: 36633286 Free PMC article.
-
Integron-sequestered dihydrofolate reductase: a recently redeployed enzyme.Trends Microbiol. 2006 May;14(5):236-42. doi: 10.1016/j.tim.2006.03.003. Epub 2006 Apr 3. Trends Microbiol. 2006. PMID: 16584884 Review.
-
Resistance to trimethoprim and sulfonamides.Vet Res. 2001 May-Aug;32(3-4):261-73. doi: 10.1051/vetres:2001123. Vet Res. 2001. PMID: 11432417 Review.
Cited by
-
Occurrence of extended- spectrum β-lactamases and ertapenem- mono- resistance in Escherichia coli isolated from diarrheal calves.One Health. 2025 Jul 10;21:101138. doi: 10.1016/j.onehlt.2025.101138. eCollection 2025 Dec. One Health. 2025. PMID: 40697494 Free PMC article.
-
Bacteria of the order Burkholderiales are original environmental hosts of type II trimethoprim resistance genes (dfrB).ISME J. 2024 Jan 8;18(1):wrae243. doi: 10.1093/ismejo/wrae243. ISME J. 2024. PMID: 39658215 Free PMC article.
-
Trimethoprim resistance in surface and wastewater is mediated by contrasting variants of the dfrB gene.ISME J. 2023 Sep;17(9):1455-1466. doi: 10.1038/s41396-023-01460-7. Epub 2023 Jun 27. ISME J. 2023. PMID: 37369703 Free PMC article.
References
-
- Tjong E., Dimri M., Mohiuddin S.S. StatPearls. StatPearls; Treasure Island, FL, USA: 2022. Biochemistry, Tetrahydrofolate. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

