Clonal Lineages and Virulence Factors of Carbapenem Resistant E. coli in Alameda County, California, 2017-2019
- PMID: 36551451
- PMCID: PMC9774732
- DOI: 10.3390/antibiotics11121794
Clonal Lineages and Virulence Factors of Carbapenem Resistant E. coli in Alameda County, California, 2017-2019
Abstract
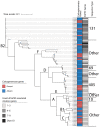
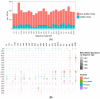
The prevalence of carbapenem-resistant Enterobacterales (CRE) has been increasing since the year 2000 and is considered a serious public health threat according to the Centers for Disease Control and Prevention. Limited studies have genotyped Carbapenem-resistant Escherichia coli using whole genome sequencing to characterize the most common lineages and resistance and virulence genes. The aim of this study was to characterize sequence data from carbapenem-resistant E. coli isolates (n = 82) collected longitudinally by the Alameda County Public Health Laboratory (ACPHL) between 2017 and 2019. E. coli genomes were screened for antibiotic resistance genes (ARGs) and extraintestinal pathogenic E. coli virulence factor genes (VFGs). The carbapenem-resistant E. coli lineages were diverse, with 24 distinct sequence types (STs) represented, including clinically important STs: ST131, ST69, ST95, and ST73. All Ambler classes of Carbapenemases were present, with NDM-5 being most the frequently detected. Nearly all isolates (90%) contained genes encoding resistance to third-generation cephalosporins; blaCTX-M genes were most common. The number of virulence genes present within pandemic STs was significantly higher than the number in non-pandemic lineages (p = 0.035). Virulence genes fimA (92%), trat (71%), kpsM (54%), and iutA (46%) were the most prevalent within the isolates. Considering the public health risk associated with CRE, these data enhance our understanding of the diversity of clinically important E. coli that are circulating in Alameda County, California.
Keywords: E. Coli; antibiotic resistance; carbapenem; genomic analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Characterization of antibiotic-susceptibility patterns and virulence genes of five major sequence types of Escherichia coli isolates cultured from extraintestinal specimens: a 1-year surveillance study from Iran.Infect Drug Resist. 2019 Apr 17;12:893-903. doi: 10.2147/IDR.S199759. eCollection 2019. Infect Drug Resist. 2019. PMID: 31118697 Free PMC article.
-
Molecular Epidemiology, Microbial Virulence, and Resistance of Carbapenem-Resistant Enterobacterales Isolates in a Teaching Hospital in Guangzhou, China.Microb Drug Resist. 2022 Jun;28(6):698-709. doi: 10.1089/mdr.2021.0156. Epub 2022 May 30. Microb Drug Resist. 2022. PMID: 35639427
-
National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain.J Antimicrob Chemother. 2011 Sep;66(9):2011-21. doi: 10.1093/jac/dkr235. Epub 2011 Jun 13. J Antimicrob Chemother. 2011. PMID: 21669946
-
Pandemic lineages of extraintestinal pathogenic Escherichia coli.Clin Microbiol Infect. 2014 May;20(5):380-90. doi: 10.1111/1469-0691.12646. Clin Microbiol Infect. 2014. PMID: 24766445 Review.
-
Bacterial characteristics of importance for recurrent urinary tract infections caused by Escherichia coli.Dan Med Bull. 2011 Apr;58(4):B4187. Dan Med Bull. 2011. PMID: 21466767 Review.
Cited by
-
Predicting sepsis mortality into an era of pandrug-resistant E. coli through modeling.Commun Med (Lond). 2024 Dec 26;4(1):278. doi: 10.1038/s43856-024-00693-7. Commun Med (Lond). 2024. PMID: 39725703 Free PMC article.
-
Carbapenem-resistant Escherichia coli exhibit diverse spatiotemporal epidemiological characteristics across the globe.Commun Biol. 2024 Jan 6;7(1):51. doi: 10.1038/s42003-023-05745-7. Commun Biol. 2024. PMID: 38184739 Free PMC article.
-
Occurrence of Imipenem-Resistant Uropathogenic Escherichia coli in Pregnant Women: An Insight into Their Virulence Profile and Clonal Structure.Curr Microbiol. 2024 Jan 9;81(2):56. doi: 10.1007/s00284-023-03576-7. Curr Microbiol. 2024. PMID: 38193903
-
Emergence of Carbapenem-Resistant Uropathogenic Escherichia coli (ST405 and ST167) Strains Carrying blaCTX-M-15, blaNDM-5 and Diverse Virulence Factors in Hospitalized Patients.Pathogens. 2024 Nov 5;13(11):964. doi: 10.3390/pathogens13110964. Pathogens. 2024. PMID: 39599517 Free PMC article.
-
High prevalence of fecal carriage of extended-spectrum beta-lactamase producing Enterobacterales among patients with urinary tract infections in rural Tanzania.Front Microbiol. 2025 Jan 6;15:1517182. doi: 10.3389/fmicb.2024.1517182. eCollection 2024. Front Microbiol. 2025. PMID: 39834365 Free PMC article.
References
-
- Rahal J.J., Urban C., Horn D., Freeman K., Segal-Maurer S., Maurer J., Mariano N., Marks S., Burns J.M., Dominick D., et al. Class Restriction of Cephalosporin Use to Control Total Cephalosporin Resistance in Nosocomial Klebsiella. JAMA. 1998;280:1233–1237. doi: 10.1001/jama.280.14.1233. - DOI - PubMed
-
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. - DOI - PubMed

