T2Bacteria and T2Resistance Assays in Critically Ill Patients with Sepsis or Septic Shock: A Descriptive Experience
- PMID: 36551480
- PMCID: PMC9774778
- DOI: 10.3390/antibiotics11121823
T2Bacteria and T2Resistance Assays in Critically Ill Patients with Sepsis or Septic Shock: A Descriptive Experience
Abstract
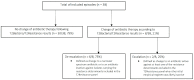
The use of rapid molecular tests may anticipate the identification of causative agents and resistance determinants in the blood of critically ill patients with sepsis. From April to December 2021, all intensive care unit patients with sepsis or septic shock who were tested with the T2Bacteria and T2Resistance assays were included in a retrospective, single center study. The primary descriptive endpoints were results of rapid molecular tests and concomitant blood cultures. Overall, 38 combinations of T2Bacteria and T2Resistance tests were performed. One or more causative agent(s) were identified by the T2Bacteria assay in 26% of episodes (10/38), whereas negative and invalid results were obtained in 66% (25/38) and 8% (3/38) of episodes, respectively. The same pathogen detected by the T2Bacteria test grew from blood cultures in 30% of cases (3/10). One or more determinant(s) of resistance were identified by the T2Resistance assay in 11% of episodes (4/38). Changes in therapy based on T2Bacteria and/or T2Resistance results occurred in 21% of episodes (8/38). In conclusion, T2Bacteria/T2Resistance results can influence early treatment decisions in critically ill patients with sepsis or septic shock in real-life practice. Large, controlled studies remain necessary to confirm a favorable impact on patients' outcomes and antimicrobial stewardship interventions.
Keywords: BSI; T2; antimicrobial resistance; diagnosis; rapid tests; sepsis; septic shock.
Conflict of interest statement
Outside the submitted work, Daniele Roberto Giacobbe reports investigator-initiated grants from Pfizer, Shionogi, and Gilead Italia and speaker and/or advisor fees from Pfizer and Tillotts Pharma. Outside the submitted work, Matteo Bassetti reports research grants and/or personal fees for advisor/consultant and/or speaker/chairman from Bayer, BioMérieux, Cidara, Cipla, Gilead, Menarini, MSD, Pfizer, and Shionogi. The other authors have no conflicts of interests to disclose.
Figures
Comment in
-
Antimicrobial stewardship at the emergency department: Dead bugs do not mutate!Eur J Intern Med. 2023 Mar;109:30-32. doi: 10.1016/j.ejim.2023.01.014. Epub 2023 Jan 19. Eur J Intern Med. 2023. PMID: 36669904 No abstract available.
References
-
- Cassini A., Hogberg L.D., Plachouras D., Quattrocchi A., Hoxha A., Simonsen G.S., Colomb-Cotinat M., Kretzschmar M.E., Devleesschauwer B., Cecchini M., et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019;19:56–66. doi: 10.1016/S1473-3099(18)30605-4. - DOI - PMC - PubMed
-
- Paul M., Carrara E., Retamar P., Tangden T., Bitterman R., Bonomo R.A., de Waele J., Daikos G.L., Akova M., Harbarth S., et al. European Society of Clinical Microbiology and Infectious Diseases (ESCMID) guidelines for the treatment of infections caused by multidrug-resistant Gram-negative bacilli (endorsed by European society of intensive care medicine) Clin. Microbiol. Infect. 2022;28:521–547. doi: 10.1016/j.cmi.2021.11.025. - DOI - PubMed
-
- Tiseo G., Brigante G., Giacobbe D.R., Maraolo A.E., Gona F., Falcone M., Giannella M., Grossi P., Pea F., Rossolini G.M., et al. Diagnosis and management of infections caused by multidrug-resistant bacteria: Guideline endorsed by the Italian Society of Infection and Tropical Diseases (SIMIT), the Italian Society of Anti-Infective Therapy (SITA), the Italian Group for Antimicrobial Stewardship (GISA), the Italian Association of Clinical Microbiologists (AMCLI) and the Italian Society of Microbiology (SIM) Int. J. Antimicrob. Agents. 2022;60:106611. doi: 10.1016/j.ijantimicag.2022.106611. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous