Single-Cell DNA Methylation Analysis in Cancer
- PMID: 36551655
- PMCID: PMC9777108
- DOI: 10.3390/cancers14246171
Single-Cell DNA Methylation Analysis in Cancer
Abstract
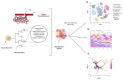
Morphological, transcriptomic, and genomic defects are well-explored parameters of cancer biology. In more recent years, the impact of epigenetic influences, such as DNA methylation, is becoming more appreciated. Aberrant DNA methylation has been implicated in many types of cancers, influencing cell type, state, transcriptional regulation, and genomic stability to name a few. Traditionally, large populations of cells from the tissue of interest are coalesced for analysis, producing averaged methylome data. Considering the inherent heterogeneity of cancer, analysing populations of cells as a whole denies the ability to discover novel aberrant methylation patterns, identify subpopulations, and trace cell lineages. Due to recent advancements in technology, it is now possible to obtain methylome data from single cells. This has both research and clinical implications, ranging from the identification of biomarkers to improved diagnostic tools. As with all emerging technologies, distinct experimental, bioinformatic, and practical challenges present themselves. This review begins with exploring the potential impact of single-cell sequencing on understanding cancer biology and how it could eventually benefit a clinical setting. Following this, the techniques and experimental approaches which made this technology possible are explored. Finally, the present challenges currently associated with single-cell DNA methylation sequencing are described.
Keywords: DNA methylation; cancer; single cell.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

