Multiplex Assay for Rapid Detection and Analysis of Nucleic Acid Using Barcode Receptor Encoded Particle (BREP)
- PMID: 36552002
- PMCID: PMC9775236
- DOI: 10.3390/biomedicines10123246
Multiplex Assay for Rapid Detection and Analysis of Nucleic Acid Using Barcode Receptor Encoded Particle (BREP)
Abstract
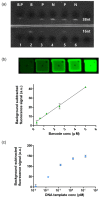
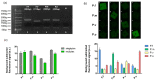
Several multiplex nucleic acid assay platforms have been developed in response to the increasing importance of nucleic acid analysis, but these assays should be optimized as per the requirements of point-of-care for clinical diagnosis. To achieve rapid and accurate detection, involving a simple procedure, we propose a new concept in the field of nucleic acid multiplex assay platforms using hydrogel microparticles, called barcode receptor-encoded particles (BREPs). The BREP assay detects multiple targets in a single reaction with a single fluorophore by analyzing graphically encoded hydrogel particles. By introducing sets of artificially synthesized barcode receptor and barcode probes, the BREP assay is easily applicable in multiplexing any genetic target; sets of barcode receptors and barcode probes should be designed delicately for universal application. The performance of the BREP assay was successfully verified in a multiplex assay for the identification of different malaria species with high sensitivity, wide dynamic range, fast detection time, and multiplexibility.
Keywords: DNA; hydrogel microparticle; molecular diagnosis; multiplex.
Conflict of interest statement
This research was utilized in the master’s degree thesis of an author (S.J.).
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

