Processing of Fluorescent Proteins May Prevent Detection of Prion Particles in [ PSI+] Cells
- PMID: 36552198
- PMCID: PMC9774836
- DOI: 10.3390/biology11121688
Processing of Fluorescent Proteins May Prevent Detection of Prion Particles in [ PSI+] Cells
Abstract
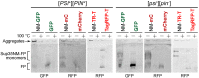
Yeast is a convenient model for studying protein aggregation as it is known to propagate amyloid prions. [PSI+] is the prion form of the release factor eRF3 (Sup35). Aggregated Sup35 causes defects in termination of translation, which results in nonsense suppression in strains carrying premature stop codons. N-terminal and middle (M) domains of Sup35 are necessary and sufficient for maintaining [PSI+] in cells while preserving the prion strain's properties. For this reason, Sup35NM fused to fluorescent proteins is often used for [PSI+] detection and investigation. However, we found that in such chimeric constructs, not all fluorescent proteins allow the reliable detection of Sup35 aggregates. Particularly, transient overproduction of Sup35NM-mCherry resulted in a diffuse fluorescent pattern in the [PSI+] cells, while no loss of prions and no effect on the Sup35NM prion properties could be observed. This effect was reproduced in various unrelated strain backgrounds and prion variants. In contrast, Sup35NM fused to another red fluorescent protein, TagRFP-T, allowed the detection of [PSI+] aggregates. Analysis of protein lysates showed that Sup35NM-mCherry is actively degraded in the cell. This degradation was not caused by vacuolar proteases and the ubiquitin-proteasomal system implicated in the Sup35 processing. Even though the intensity of this proteolysis was higher than that of Sup35NM-GFP, it was roughly the same as in the case of Sup35NM-TagRFP-T. Thus, it is possible that, in contrast to TagRFP-T, degradation products of Sup35NM-mCherry still preserve their fluorescent properties while losing the ability to decorate pre-existing Sup35 aggregates. This results in diffuse fluorescence despite the presence of the prion aggregates in the cell. Thus, tagging with fluorescent proteins should be used with caution, as such proteolysis may increase the rate of false-negative results when detecting prion-bearing cells.
Keywords: GFP; RFP; Sup35; [PSI+]; mCherry; prion; yeast.
Conflict of interest statement
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





Similar articles
-
Yeast prion protein New1 can break Sup35 amyloid fibrils into fragments in an ATP-dependent manner.Genes Cells. 2011 May;16(5):545-56. doi: 10.1111/j.1365-2443.2011.01510.x. Epub 2011 Apr 1. Genes Cells. 2011. PMID: 21453424
-
Interaction of human laminin receptor with Sup35, the [PSI⁺] prion-forming protein from S. cerevisiae: a yeast model for studies of LamR interactions with amyloidogenic proteins.PLoS One. 2014 Jan 8;9(1):e86013. doi: 10.1371/journal.pone.0086013. eCollection 2014. PLoS One. 2014. PMID: 24416454 Free PMC article.
-
Q-Rich Yeast Prion [PSI+] Accelerates Aggregation of Transthyretin, a Non-Q-Rich Human Protein.Front Mol Neurosci. 2018 Mar 13;11:75. doi: 10.3389/fnmol.2018.00075. eCollection 2018. Front Mol Neurosci. 2018. PMID: 29593496 Free PMC article.
-
A bipolar personality of yeast prion proteins.Prion. 2011 Oct-Dec;5(4):305-10. doi: 10.4161/pri.18307. Epub 2011 Oct 1. Prion. 2011. PMID: 22156730 Free PMC article. Review.
-
Sup35 methionine oxidation is a trigger for de novo [PSI(+)] prion formation.Prion. 2015;9(4):257-65. doi: 10.1080/19336896.2015.1065372. Prion. 2015. PMID: 26267336 Free PMC article. Review.
Cited by
-
Intracellular Degradation of SARS-CoV-2 N-Protein Caused by Modular Nanotransporters Containing Anti-N-Protein Monobody and a Sequence That Recruits the Keap1 E3 Ligase.Pharmaceutics. 2023 Dec 19;16(1):4. doi: 10.3390/pharmaceutics16010004. Pharmaceutics. 2023. PMID: 38276482 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

