Rice TCD8 Encoding a Multi-Domain GTPase Is Crucial for Chloroplast Development of Early Leaf Stage at Low Temperatures
- PMID: 36552248
- PMCID: PMC9774597
- DOI: 10.3390/biology11121738
Rice TCD8 Encoding a Multi-Domain GTPase Is Crucial for Chloroplast Development of Early Leaf Stage at Low Temperatures
Abstract
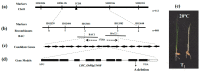
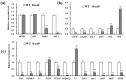
The multi-domain GTPase (MnmE) is conservative from bacteria to human and participates in tRNA modified synthesis. However, our understanding of how the MnmE is involved in plant chloroplast development is scarce, let alone in rice. A novel rice mutant, thermo-sensitive chlorophyll-deficient mutant 8 (tcd8) was identified in this study, which apparently presented an albino phenotype at 20 °C but a normal green over 24 °C, coincided with chloroplast development and chlorophyll content. Map-based cloning and complementary test revealed the TCD8 encoded a multi-domain GTPase localized in chloroplasts. In addition, the disturbance of TCD8 suppressed the transcripts of certain chloroplast-related genes at low temperature, although the genes were recoverable to nearly normal levels at high temperature (32 °C), indicating that TCD8 governs chloroplast development at low temperature. The multi-domain GTPase gene in rice is first reported in this study, which endorses the importance in exploring chloroplast development in rice.
Keywords: albino phenotype; chloroplast; low temperature; multi-domain GTPase; rice.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Importance of the rice TCD9 encoding α subunit of chaperonin protein 60 (Cpn60α) for the chloroplast development during the early leaf stage.Plant Sci. 2014 Feb;215-216:172-9. doi: 10.1016/j.plantsci.2013.11.003. Epub 2013 Nov 18. Plant Sci. 2014. PMID: 24388528
-
Rice TSV3 Encoding Obg-Like GTPase Protein Is Essential for Chloroplast Development During the Early Leaf Stage Under Cold Stress.G3 (Bethesda). 2018 Jan 4;8(1):253-263. doi: 10.1534/g3.117.300249. G3 (Bethesda). 2018. PMID: 29162684 Free PMC article.
-
Rice TSV2 encoding threonyl-tRNA synthetase is needed for early chloroplast development and seedling growth under cold stress.G3 (Bethesda). 2021 Sep 6;11(9):jkab196. doi: 10.1093/g3journal/jkab196. G3 (Bethesda). 2021. PMID: 34544147 Free PMC article.
-
The Rice TCM5 Gene Encoding a Novel Deg Protease Protein is Essential for Chloroplast Development under High Temperatures.Rice (N Y). 2016 Dec;9(1):13. doi: 10.1186/s12284-016-0086-5. Epub 2016 Mar 21. Rice (N Y). 2016. PMID: 27000876 Free PMC article.
-
Establishment of the chloroplast genetic system in rice during early leaf development and at low temperatures.Front Plant Sci. 2014 Aug 11;5:386. doi: 10.3389/fpls.2014.00386. eCollection 2014. Front Plant Sci. 2014. PMID: 25157260 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

