A Systems Biology and LASSO-Based Approach to Decipher the Transcriptome-Interactome Signature for Predicting Non-Small Cell Lung Cancer
- PMID: 36552262
- PMCID: PMC9774707
- DOI: 10.3390/biology11121752
A Systems Biology and LASSO-Based Approach to Decipher the Transcriptome-Interactome Signature for Predicting Non-Small Cell Lung Cancer
Abstract
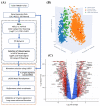
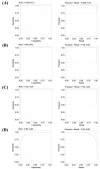
The lack of precise molecular signatures limits the early diagnosis of non-small cell lung cancer (NSCLC). The present study used gene expression data and interaction networks to develop a highly accurate model with the least absolute shrinkage and selection operator (LASSO) for predicting NSCLC. The differentially expressed genes (DEGs) were identified in NSCLC compared with normal tissues using TCGA and GTEx data. A biological network was constructed using DEGs, and the top 20 upregulated and 20 downregulated hub genes were identified. These hub genes were used to identify signature genes with penalized logistic regression using the LASSO to predict NSCLC. Our model’s development involved the following steps: (i) the dataset was divided into 80% for training (TR) and 20% for testing (TD1); (ii) a LASSO logistic regression analysis was performed on the TR with 10-fold cross-validation and identified a combination of 17 genes as NSCLC predictors, which were used further for development of the LASSO model. The model’s performance was assessed on the TD1 dataset and achieved an accuracy and an area under the curve of the receiver operating characteristics (AUC-ROC) of 0.986 and 0.998, respectively. Furthermore, the performance of the LASSO model was evaluated using three independent NSCLC test datasets (GSE18842, GSE27262, GSE19804) and achieved high accuracy, with an AUC-ROC of >0.99, >0.99, and 0.95, respectively. Based on this study, a web application called NSCLCpred was developed to predict NSCLC.
Keywords: LASSO model; artificial intelligence; biological networks; gene expression; hub genes; non-small cell lung cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Walker M.S., Wong W., Ravelo A., Miller P.J.E., Schwartzberg L.S. Effectiveness outcomes and health related quality of life impact of disease progression in patients with advanced nonsquamous NSCLC treated in real-world community oncology settings: Results from a prospective medical record registry study. Health Qual. Life Outcomes. 2017;15:160. doi: 10.1186/s12955-017-0735-4. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

