Advances in Production of Hydroxycinnamoyl-Quinic Acids: From Natural Sources to Biotechnology
- PMID: 36552635
- PMCID: PMC9774772
- DOI: 10.3390/antiox11122427
Advances in Production of Hydroxycinnamoyl-Quinic Acids: From Natural Sources to Biotechnology
Abstract
Hydroxycinnamoyl-quinic acids (HCQAs) are polyphenol esters formed of hydroxycinnamic acids and (-)-quinic acid. They are naturally synthesized by plants and some micro-organisms. The ester of caffeic acid and quinic acid, the chlorogenic acid, is an intermediate of lignin biosynthesis. HCQAs are biologically active dietary compounds exhibiting several important therapeutic properties, including antioxidant, antimicrobial, anti-inflammatory, neuroprotective, and other activities. They can also be used in the synthesis of nanoparticles or drugs. However, extraction of these compounds from biomass is a complex process and their synthesis requires costly precursors, limiting the industrial production and availability of a wider variety of HCQAs. The recently emerged production through the bioconversion is still in an early stage of development. In this paper, we discuss existing and potential future strategies for production of HCQAs.
Keywords: antioxidants; biosynthesis by engineered micro-organisms; chlorogenic acid; extraction; hydroxycinnamoyl-quinic acids; synthesis.
Conflict of interest statement
The authors declare no conflict of interest. The funding source had no role in the study’s design; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



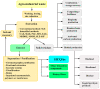
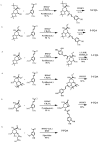


References
-
- DataIntelo Global Chlorogenic Acid Market Report, History and Forecast 2014–2025, Breakdown Data by Manufacturers, Key Regions, Types and Application. [(accessed on 15 March 2022)]. Available online: https://dataintelo.com/report/chlorogenic-acid-market.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

