PICALM and Alzheimer's Disease: An Update and Perspectives
- PMID: 36552756
- PMCID: PMC9776874
- DOI: 10.3390/cells11243994
PICALM and Alzheimer's Disease: An Update and Perspectives
Abstract
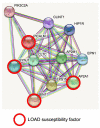
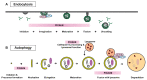
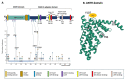
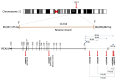
Genome-wide association studies (GWAS) have identified the PICALM (Phosphatidylinositol binding clathrin-assembly protein) gene as the most significant genetic susceptibility locus after APOE and BIN1. PICALM is a clathrin-adaptor protein that plays a critical role in clathrin-mediated endocytosis and autophagy. Since the effects of genetic variants of PICALM as AD-susceptibility loci have been confirmed by independent genetic studies in several distinct cohorts, there has been a number of in vitro and in vivo studies attempting to elucidate the underlying mechanism by which PICALM modulates AD risk. While differential modulation of APP processing and Aβ transcytosis by PICALM has been reported, significant effects of PICALM modulation of tau pathology progression have also been evidenced in Alzheimer's disease models. In this review, we summarize the current knowledge about PICALM, its physiological functions, genetic variants, post-translational modifications and relevance to AD pathogenesis.
Keywords: Alzheimer’s disease; GWAS; PICALM; amyloid β; microglia; neurofibrillary tangles.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

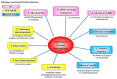
References
-
- Brion J.P., Couck A.M., Passareiro E., Flament-Durand J. Neurofibrillary Tangles of Alzheimer’s Disease: An Immunohistochemical Study. J. Submicrosc. Cytol. 1985;17:89–96. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

