Gain- and Loss-of-Function CFTR Alleles Are Associated with COVID-19 Clinical Outcomes
- PMID: 36552859
- PMCID: PMC9776607
- DOI: 10.3390/cells11244096
Gain- and Loss-of-Function CFTR Alleles Are Associated with COVID-19 Clinical Outcomes
Abstract
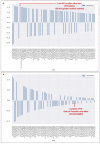
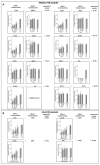
Carriers of single pathogenic variants of the CFTR (cystic fibrosis transmembrane conductance regulator) gene have a higher risk of severe COVID-19 and 14-day death. The machine learning post-Mendelian model pinpointed CFTR as a bidirectional modulator of COVID-19 outcomes. Here, we demonstrate that the rare complex allele [G576V;R668C] is associated with a milder disease via a gain-of-function mechanism. Conversely, CFTR ultra-rare alleles with reduced function are associated with disease severity either alone (dominant disorder) or with another hypomorphic allele in the second chromosome (recessive disorder) with a global residual CFTR activity between 50 to 91%. Furthermore, we characterized novel CFTR complex alleles, including [A238V;F508del], [R74W;D1270N;V201M], [I1027T;F508del], [I506V;D1168G], and simple alleles, including R347C, F1052V, Y625N, I328V, K68E, A309D, A252T, G542*, V562I, R1066H, I506V, I807M, which lead to a reduced CFTR function and thus, to more severe COVID-19. In conclusion, CFTR genetic analysis is an important tool in identifying patients at risk of severe COVID-19.
Keywords: CFTR complex alleles; COVID-19; post-Mendelian model.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., et al. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. - DOI - PMC - PubMed
-
- Grasselli G., Greco M., Zanella A., Albano G., Antonelli M., Bellani G., Bonanomi E., Cabrini L., Carlesso E., Castelli G., et al. Risk Factors Associated With Mortality Among Patients With COVID-19 in Intensive Care Units in Lombardy, Italy. JAMA Intern. Med. 2020;180:1345–1355. doi: 10.1001/jamainternmed.2020.3539. - DOI - PMC - PubMed
-
- Severe COVID-19 GWAS Group. Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., Fernández J., Prati D., Baselli G., et al. Genomewide Association Study of Severe COVID-19 with Respiratory Failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/nejmoa2020283. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

