Predicting IDH Mutation Status in Low-Grade Gliomas Based on Optimal Radiomic Features Combined with Multi-Sequence Magnetic Resonance Imaging
- PMID: 36553002
- PMCID: PMC9776893
- DOI: 10.3390/diagnostics12122995
Predicting IDH Mutation Status in Low-Grade Gliomas Based on Optimal Radiomic Features Combined with Multi-Sequence Magnetic Resonance Imaging
Abstract
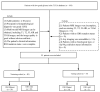
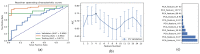
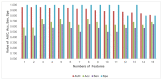
The IDH somatic mutation status is an important basis for the diagnosis and classification of gliomas. We proposed a "6-Step" general radiomics model to noninvasively predict the IDH mutation status by simultaneously tuning combined multi-sequence MRI and optimizing the full radiomics processing pipeline. Radiomic features (n = 3776) were extracted from multi-sequence MRI (T1, T2, FLAIR, and T1Gd) in low-grade gliomas (LGGs), and a total of 45,360 radiomics pipeline were investigated according to different settings. The predictive ability of the general radiomics model was evaluated with regards to accuracy, stability, and efficiency. Based on numerous experiments, we finally reached an optimal pipeline for classifying IDH mutation status, namely the T2+FLAIR combined multi-sequence with the wavelet image filter, mean data normalization, PCC dimension reduction, RFE feature selection, and SVM classifier. The mean and standard deviation of AUC, accuracy, sensitivity, and specificity were 0.873 ± 0.05, 0.876 ± 0.09, 0.875 ± 0.11, and 0.877 ± 0.15, respectively. Furthermore, 14 radiomic features that best distinguished the IDH mutation status of the T2+FLAIR multi-sequence were analyzed, and the gray level co-occurrence matrix (GLCM) features were shown to be of high importance. Apart from the promising prediction of the molecular subtypes, this study also provided a general tool for radiomics investigation.
Keywords: IDH; glioma; machine learning; multi-sequence MRI; radiomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Wang L.-B., Karpova A., Gritsenko M.A., Kyle J.E., Cao S., Li Y., Rykunov D., Colaprico A., Rothstein J.H., Hong R., et al. Clinical Proteomic Tumor Analysis Consortium. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell. 2021;39:509–528. doi: 10.1016/j.ccell.2021.01.006. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

