A Multigraph-Based Representation of Hi-C Data
- PMID: 36553456
- PMCID: PMC9778156
- DOI: 10.3390/genes13122189
A Multigraph-Based Representation of Hi-C Data
Abstract
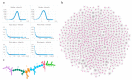
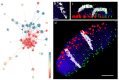
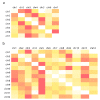
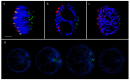
Chromatin-chromatin interactions and three-dimensional (3D) spatial structures are involved in transcriptional regulation and have a decisive role in DNA replication and repair. To understand how individual genes and their regulatory elements function within the larger genomic context, and how the genome reacts to environmental stimuli, the linear sequence information needs to be interpreted in three-dimensional space, which is still a challenging task. Here, we propose a novel, heuristic approach to represent Hi-C datasets by a whole-genomic pseudo-structure in 3D space. The baseline of our approach is the construction of a multigraph from genomic-sequence data and Hi-C interaction data, then applying a modified force-directed layout algorithm. The resulting layout is a pseudo-structure. While pseudo-structures are not based on direct observation and their details are inherent to settings, surprisingly, they demonstrate interesting, overall similarities of known genome structures of both barley and rice, namely, the Rabl and Rosette-like conformation. It has an exciting potential to be extended by additional omics data (RNA-seq, Chip-seq, etc.), allowing to visualize the dynamics of the pseudo-structures across various tissues or developmental stages. Furthermore, this novel method would make it possible to revisit most Hi-C data accumulated in the public domain in the last decade.
Keywords: DNA; Hi-C; barley; graph theory; rice.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

