Dbf4 Zn-Finger Motif Is Specifically Required for Stimulation of Ctf19-Activated Origins in Saccharomyces cerevisiae
- PMID: 36553469
- PMCID: PMC9778208
- DOI: 10.3390/genes13122202
Dbf4 Zn-Finger Motif Is Specifically Required for Stimulation of Ctf19-Activated Origins in Saccharomyces cerevisiae
Abstract
Eukaryotic genomes are replicated in spatiotemporal patterns that are stereotypical for individual genomes and developmental profiles. In the model system Saccharomyces cerevisiae, two primary mechanisms determine the preferential activation of replication origins during early S phase, thereby largely defining the consequent replication profiles of these cells. Both mechanisms are thought to act through specific recruitment of a rate-limiting initiation factor, Dbf4-dependent kinase (DDK), to a subset of licensed replication origins. Fkh1/2 is responsible for stimulation of most early-firing origins, except for centromere (CEN)-proximal origins that recruit DDK via the kinetochore protein Ctf19, which is required for their early firing. The C-terminus of Dbf4 has been implicated in its recruitment to origins via both the Fkh1/2 and Ctf19 mechanisms. Here, we show that the Zn-finger motif within the C-terminus is specifically required for Dbf4 recruitment to CENs to stimulate CEN-proximal/Ctf19-dependent origins, whereas stimulation of origins via the Fkh1/2 pathway remains largely intact. These findings re-open the question of exactly how Fkh1/2 and DDK act together to stimulate replication origin initiation.
Keywords: Ctf19; Dbf4-dependent-kinase; Fkh1; chromosome domains; replication origins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

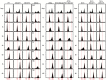

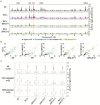
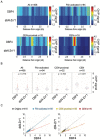
References
-
- Natsume T., Muller C.A., Katou Y., Retkute R., Gierlinski M., Araki H., Blow J.J., Shirahige K., Nieduszynski C.A., Tanaka T.U. Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment. Mol. Cell. 2013;50:661–674. doi: 10.1016/j.molcel.2013.05.011. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

