The Identification of Large Rearrangements Involving Intron 2 of the CDH1 Gene in BRCA1/2 Negative and Breast Cancer Susceptibility
- PMID: 36553480
- PMCID: PMC9778491
- DOI: 10.3390/genes13122213
The Identification of Large Rearrangements Involving Intron 2 of the CDH1 Gene in BRCA1/2 Negative and Breast Cancer Susceptibility
Abstract
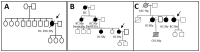
E-cadherin, a CDH1 gene product, is a calcium-dependent cell-cell adhesion molecule playing a critical role in the establishment of epithelial architecture, maintenance of cell polarity, and differentiation. Germline pathogenic variants in the CDH1 gene are associated with hereditary diffuse gastric cancer (HDGC), and large rearrangements in the CDH1 gene are now being reported as well. Because CDH1 pathogenic variants could be associated with breast cancer (BC) susceptibility, CDH1 rearrangements could also impact it. The aim of our study is to identify rearrangements in the CDH1 gene in 148 BC cases with no BRCA1 and BRCA2 pathogenic variants. To do so, a zoom-in CGH array, covering the exonic, intronic, and flanking regions of the CDH1 gene, was used to screen our cohort. Intron 2 of the CDH1 gene was specifically targeted because it is largely reported to include several regulatory regions. As results, we detected one large rearrangement causing a premature stop in exon 3 of the CDH1 gene in a proband with a bilateral lobular breast carcinoma and a gastric carcinoma (GC). Two large rearrangements in the intron 2, a deletion and a duplication, were also reported only with BC cases without any familial history of GC. No germline rearrangements in the CDH1 coding region were detected in those families without GC and with a broad range of BC susceptibility. This study confirms the diversity of large rearrangements in the CDH1 gene. The rearrangements identified in intron 2 highlight the putative role of this intron in CDH1 regulation and alternative transcripts. Recurrent duplication copy number variations (CNV) are found in this region, and the deletion encompasses an alternative CDH1 transcript. Screening for large rearrangements in the CDH1 gene could be important for genetic testing of BC.
Keywords: BRCA1/2 negative cases; CDH1 rearrangements; CGH array; CNV; breast carcinoma; gastric carcinoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Rostami P., Zendehdel K., Shirkoohi R., Ebrahimi E., Ataei M., Imanian H., Najmabadi H., Akbari M.R., Sanati M.H. Gene Panel Testing in Hereditary Breast Cancer. Arch. Iran. Med. Acad. Med. Sci. IR Iran. 2020;23:155–162. - PubMed
-
- Ford D., Easton D.F., Stratton M., Narod S., Goldgar D., Devilee P., Bishop D.T., Weber B., Lenoir G., Chang-Claude J., et al. Genetic Heterogeneity and Penetrance Analysis of the BRCA1 and BRCA2 Genes in Breast Cancer Families. Am. J. Hum. Genet. 1998;62:676–689. doi: 10.1086/301749. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

