Characterising Mitochondrial Capture in an Iberian Shrew
- PMID: 36553495
- PMCID: PMC9777731
- DOI: 10.3390/genes13122228
Characterising Mitochondrial Capture in an Iberian Shrew
Abstract
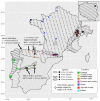
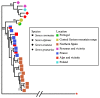
Mitochondrial introgression raises questions of biogeography and of the extent of reproductive isolation and natural selection. Previous phylogenetic work on the Sorex araneus complex revealed apparent mitonuclear discordance in Iberian shrews, indicating past hybridisation of Sorex granarius and the Carlit chromosomal race of S. araneus, enabling introgression of the S. araneus mitochondrial genome into S. granarius. To further study this, we genetically typed 61 Sorex araneus/coronatus/granarius from localities in Portugal, Spain, France, and Andorra at mitochondrial, autosomal, and sex-linked loci and combined our data with the previously published sequences. Our data are consistent with earlier data indicating that S. coronatus and S. granarius are the most closely related of the three species, confirming that S. granarius from the Central System mountain range in Spain captured the mitochondrial genome from a population of S. araneus. This mitochondrial capture event can be explained by invoking a biogeographical scenario whereby S. araneus was in contact with S. granarius during the Younger Dryas in central Iberia, despite the two species currently having disjunct distributions. We discuss whether selection favoured S. granarius with an introgressed mitochondrial genome. Our data also suggest recent hybridisation and introgression between S. coronatus and S. granarius, as well as between S. araneus and S. coronatus.
Keywords: Iberia; Sorex araneus complex; hybridisation; introgression; karyotype; phylogenetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Moritz C., Dowling T.E., Brown W.M. Evolution of animal mitochondrial DNA: Relevance for population biology and systematics. Ann. Rev. Ecol. Syst. 1987;18:269–292. doi: 10.1146/annurev.es.18.110187.001413. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

