In Vitro Analysis of Biological Activity of Circulating Cell-Free DNA Isolated from Blood Plasma of Schizophrenic Patients and Healthy Controls-Part 2: Adaptive Response
- PMID: 36553550
- PMCID: PMC9777734
- DOI: 10.3390/genes13122283
In Vitro Analysis of Biological Activity of Circulating Cell-Free DNA Isolated from Blood Plasma of Schizophrenic Patients and Healthy Controls-Part 2: Adaptive Response
Abstract
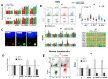
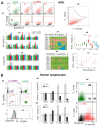
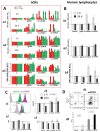
Oxidized in vitro genomic DNA (gDNA) is known to launch an adaptive response in human cell cultures. The cfDNA extracted from the plasma of schizophrenic patients (sz-cfDNA) and healthy controls (hc-cfDNA) contains increased amounts of 8-oxodG, a DNA-oxidation marker. The aim of the research was answering a question: can the human cfDNA isolated from blood plasma stimulate the adaptive response in human cells? In vitro responses of ten human skin fibroblasts (HSFs) and four peripheral blood mononuclear cell (PBMC) lines after 1-24 h of incubation with sz-cfDNA, gDNA and hc-cfDNA containing different amounts of 8-oxodG were examined. Expressions of RNA of eight genes (NOX4, NFE2L2, SOD1, HIF1A, BRCA1, BRCA2, BAX and BCL2), six proteins (NOX4, NRF2, SOD1, HIF1A, γH2AX and BRCA1) and DNA-oxidation marker 8-oxodG were analyzed by RT-qPCR and flow cytometry (when analyzing the data, a subpopulation of lymphocytes (PBL) was identified). Adding hc-cfDNA or sz-cfDNA to HSFs or PBMC media in equal amounts (50 ng/mL, 1-3 h) stimulated transient synthesis of free radicals (ROS), which correlated with an increase in the expressions of NOX4 and SOD1 genes and with an increase in the levels of the markers of DNA damage γH2AX and 8-oxodG. ROS and DNA damage induced an antioxidant response (expression of NFE2L2 and HIF1A), DNA damage response (BRCA1 and BRCA2 gene expression) and anti-apoptotic response (changes in BAX and BCL2 genes expression). Heterogeneity of cells of the same HSFs or PBL population was found with respect to the type of response to (sz,hc)-cfDNA. Most cells responded to oxidative stress with an increase in the amount of NRF2 and BRCA1 proteins along with a moderate increase in the amount of NOX4 protein and a low amount of 8-oxodG oxidation marker. However, upon the exposure to (sz,hc)-cfDNA, the size of the subpopulation with apoptosis signs (high DNA damage degree, high NOX4 and low NRF2 and BRCA1 levels) also increased. No significant difference between the responses to sz-cfDNA and hc-cfDNA was observed. Sz-cfDNA and hc-cfDNA showed similarly high bioactivity towards fibroblasts and lymphocytes. Conclusion: In cultured human cells, hc-cfDNA and sz-cfDNA equally stimulated an adaptive response aimed at launching the antioxidant, repair, and anti-apoptotic processes. The mediator of the development of the adaptive response are ROS produced by, among others, NOX4 and SOD1 enzymes.
Keywords: BRCA1; HIF1A; NOX4; NRF2; SOD1; adaptive response; cell-free DNA; schizophrenia; γH2AX.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

