Pantothenate and L-Carnitine Supplementation Improves Pathological Alterations in Cellular Models of KAT6A Syndrome
- PMID: 36553567
- PMCID: PMC9778406
- DOI: 10.3390/genes13122300
Pantothenate and L-Carnitine Supplementation Improves Pathological Alterations in Cellular Models of KAT6A Syndrome
Abstract
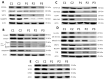
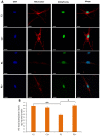
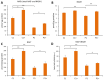
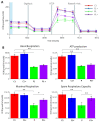
Mutations in several genes involved in the epigenetic regulation of gene expression have been considered risk alterations to different intellectual disability (ID) syndromes associated with features of autism spectrum disorder (ASD). Among them are the pathogenic variants of the lysine-acetyltransferase 6A (KAT6A) gene, which causes KAT6A syndrome. The KAT6A enzyme participates in a wide range of critical cellular functions, such as chromatin remodeling, gene expression, protein synthesis, cell metabolism, and replication. In this manuscript, we examined the pathophysiological alterations in fibroblasts derived from three patients harboring KAT6A mutations. We addressed survival in a stress medium, histone acetylation, protein expression patterns, and transcriptome analysis, as well as cell bioenergetics. In addition, we evaluated the therapeutic effectiveness of epigenetic modulators and mitochondrial boosting agents, such as pantothenate and L-carnitine, in correcting the mutant phenotype. Pantothenate and L-carnitine treatment increased histone acetylation and partially corrected protein and transcriptomic expression patterns in mutant KAT6A cells. Furthermore, the cell bioenergetics of mutant cells was significantly improved. Our results suggest that pantothenate and L-carnitine can significantly improve the mutant phenotype in cellular models of KAT6A syndrome.
Keywords: KAT6A syndrome; L-carnitine; histone acetylation; intellectual disability; lysine acetyltransferase 6 A; pantothenate.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Arboleda V.A., Lee H., Dorrani N., Zadeh N., Willis M., Macmurdo C.F., Manning M.A., Kwan A., Hudgins L., Barthelemy F., et al. De novo nonsense mutations in KAT6A, a lysine acetyl-transferase gene, cause a syndrome including microcephaly and global developmental delay. Am. J. Hum. Genet. 2015;96:498–506. doi: 10.1016/j.ajhg.2015.01.017. - DOI - PMC - PubMed
-
- Tham E., Lindstrand A., Santani A., Malmgren H., Nesbitt A., Dubbs H.A., Zackai E.H., Parker M.J., Millan F., Rosenbaum K., et al. Dominant mutations in KAT6A cause intellectual disability with recognizable syndromic features. Am. J. Hum. Genet. 2015;96:507–513. doi: 10.1016/j.ajhg.2015.01.016. - DOI - PMC - PubMed
-
- Millan F., Cho M.T., Retterer K., Monaghan K.G., Bai R., Vitazka P., Everman D.B., Smith B., Angle B., Roberts V., et al. Whole exome sequencing reveals de novo pathogenic variants in KAT6A as a cause of a neurodevelopmental disorder. Am. J. Med. Genet. A. 2016;170:1791–1798. doi: 10.1002/ajmg.a.37670. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

