Effects of N-Acetylcysteine on the Proliferation, Hormone Secretion Level, and Gene Expression Profiles of Goat Ovarian Granulosa Cells
- PMID: 36553574
- PMCID: PMC9778279
- DOI: 10.3390/genes13122306
Effects of N-Acetylcysteine on the Proliferation, Hormone Secretion Level, and Gene Expression Profiles of Goat Ovarian Granulosa Cells
Abstract
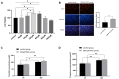
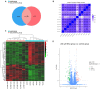
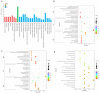
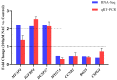
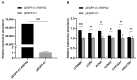
The purpose of this paper was to investigate the effects of N-acetylcysteine (NAC) on the proliferation, hormone secretion, and mRNA expression profiles of ovarian granulosa cells (GCs) in vitro. A total of 12 ovaries from 6 follicular-stage goats were collected for granulosa cell extraction. The optimum concentration of NAC addition was determined to be 200 μM via the Cell Counting Kit 8 (CCK-8) method. Next, GCs were cultured in a medium supplemented with 200 μM NAC (200 μM NAC group) and 0 μ M NAC (control group) for 48 h. The effects of 200 μM NAC on the proliferation of granulosa cells and hormones were studied by 5-ethynyl-2'-deoxyuridine (EdU) assay and enzyme-linked immunosorbent assay (ELISA). mRNA expression was analyzed by transcriptome sequencing. The results indicate that 200 μM NAC significantly increased cell viability and the proportion of cells in the S phase but promoted hormone secretion to a lesser degree. Overall, 122 differentially expressed genes (DEGs) were identified. A total of 51 upregulated and 71 downregulated genes were included. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses indicated that the most DEGs were enriched in terms of cell growth regulation, cell growth, neuroactive ligand-receptor interaction, cytokine-cytokine receptor interaction, the cAMP-signaling pathway, and the Wnt-signaling pathway. Seven genes related to granulosa cell proliferation were screened, IGFBP4, HTRA4, SST, SSTR1, WISP1, DAAM2, and RSPO2. The above results provide molecular theoretical support for NAC as a feed additive to improve follicle development and improve reproductive performance in ewes.
Keywords: N-acetylcysteine; RNA-seq; mRNA expression; ovarian granulosa cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

