Deviations from Mendelian Inheritance on Bovine X-Chromosome Revealing Recombination, Sex-of-Offspring Effects and Fertility-Related Candidate Genes
- PMID: 36553588
- PMCID: PMC9778079
- DOI: 10.3390/genes13122322
Deviations from Mendelian Inheritance on Bovine X-Chromosome Revealing Recombination, Sex-of-Offspring Effects and Fertility-Related Candidate Genes
Abstract
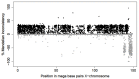
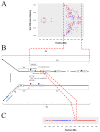
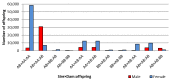
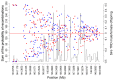
Transmission ratio distortion (TRD), or significant deviations from Mendelian inheritance, is a well-studied phenomenon on autosomal chromosomes, but has not yet received attention on sex chromosomes. TRD was analyzed on 3832 heterosomal single nucleotide polymorphisms (SNPs) and 400 pseudoautosomal SNPs spanning the length of the X-chromosome using 436,651 genotyped Holstein cattle. On the pseudoautosomal region, an opposite sire-TRD pattern between male and female offspring was identified for 149 SNPs. This finding revealed unique SNPs linked to a specific-sex (Y- or X-) chromosome and describes the accumulation of recombination events across the pseudoautosomal region. On the heterosomal region, 13 SNPs and 69 haplotype windows were identified with dam-TRD. Functional analyses for TRD regions highlighted relevant biological functions responsible to regulate spermatogenesis, development of Sertoli cells, homeostasis of endometrium tissue and embryonic development. This study uncovered the prevalence of different TRD patterns across both heterosomal and pseudoautosomal regions of the X-chromosome and revealed functional candidate genes for bovine reproduction.
Keywords: bovine; chromosome X; recombination; transmission ratio distortion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Casellas J., Cañas-Álvarez J.J., González-Rodríguez A., Puig-Oliveras A., Fina M., Piedrafita J., Molina A., Díaz C., Baró J.A., Varona L. Bayesian Analysis of Parent-Specific Transmission Ratio Distortion in Seven Spanish Beef Cattle Breeds. Anim. Genet. 2017;48:93–96. doi: 10.1111/age.12509. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

