Differential Allele-Specific Expression Revealed Functional Variants and Candidate Genes Related to Meat Quality Traits in B. indicus Muscle
- PMID: 36553605
- PMCID: PMC9777870
- DOI: 10.3390/genes13122336
Differential Allele-Specific Expression Revealed Functional Variants and Candidate Genes Related to Meat Quality Traits in B. indicus Muscle
Abstract
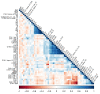
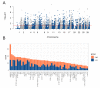
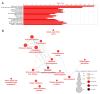
Traditional transcriptomics approaches have been used to identify candidate genes affecting economically important livestock traits. Regulatory variants affecting these traits, however, remain under covered. Genomic regions showing allele-specific expression (ASE) are under the effect of cis-regulatory variants, being useful for improving the accuracy of genomic selection models. Taking advantage of the better of these two methods, we investigated single nucleotide polymorphisms (SNPs) in regions showing differential ASE (DASE SNPs) between contrasting groups for beef quality traits. For these analyses, we used RNA sequencing data, imputed genotypes and genomic estimated breeding values of muscle-related traits from 190 Nelore (Bos indicus) steers. We selected 40 contrasting unrelated samples for the analysis (N = 20 animals per contrasting group) and used a beta-binomial model to identify ASE SNPs in only one group (i.e., DASE SNPs). We found 1479 DASE SNPs (FDR ≤ 0.05) associated with 55 beef-quality traits. Most DASE genes were involved with tenderness and muscle homeostasis, presenting a co-expression module enriched for the protein ubiquitination process. The results overlapped with epigenetics and phenotype-associated data, suggesting that DASE SNPs are potentially linked to cis-regulatory variants affecting simultaneously the transcription and phenotype through chromatin state modulation.
Keywords: ASE; beef; cis-regulation; phenotype.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Huang W.-C., Ferris E., Cheng T., Hörndli C.S., Gleason K., Tamminga C., Wagner J.D., Boucher K.M., Christian J.L., Gregg C. Diverse Non-Genetic, Allele-Specific Expression Effects Shape Genetic Architecture at the Cellular Level in the Mammalian Brain. Neuron. 2017;93:1094–1109.e7. doi: 10.1016/j.neuron.2017.01.033. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

