Genome-Wide Association Studies in Sunflower: Towards Sclerotinia sclerotiorum and Diaporthe/Phomopsis Resistance Breeding
- PMID: 36553624
- PMCID: PMC9777803
- DOI: 10.3390/genes13122357
Genome-Wide Association Studies in Sunflower: Towards Sclerotinia sclerotiorum and Diaporthe/Phomopsis Resistance Breeding
Abstract
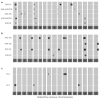
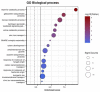
Diseases caused by necrotrophic fungi, such as the cosmopolitan Sclerotinia sclerotiorum and the Diaporthe/Phomopsis complex, are among the most destructive diseases of sunflower worldwide. The lack of complete resistance combined with the inefficiency of chemical control makes assisted breeding the best strategy for disease control. In this work, we present an integrated genome-wide association (GWA) study investigating the response of a diverse panel of sunflower inbred lines to both pathogens. Phenotypic data for Sclerotinia head rot (SHR) consisted of five disease descriptors (disease incidence, DI; disease severity, DS; area under the disease progress curve for DI, AUDPCI, and DS, AUDPCS; and incubation period, IP). Two disease descriptors (DI and DS) were evaluated for two manifestations of Diaporthe/Phomopsis: Phomopsis stem canker (PSC) and Phomopsis head rot (PHR). In addition, a principal component (PC) analysis was used to derive transformed phenotypes as inputs to a univariate GWA (PC-GWA). Genotypic data comprised a panel of 4269 single nucleotide polymorphisms (SNP), generated via genotyping-by-sequencing. The GWA analysis revealed 24 unique marker-trait associations for SHR, 19 unique marker-trait associations for Diaporthe/Phomopsis diseases, and 7 markers associated with PC1 and PC2. No common markers were found for the response to the two pathogens. Nevertheless, epistatic interactions were identified between markers significantly associated with the response to S. sclerotiorum and Diaporthe/Phomopsis. This suggests that, while the main determinants of resistance may differ for the two pathogens, there could be an underlying common genetic basis. The exploration of regions physically close to the associated markers yielded 364 genes, of which 19 were predicted as putative disease resistance genes. This work presents the first simultaneous evaluation of two manifestations of Diaporthe/Phomopsis in sunflower, and undertakes a comprehensive GWA study by integrating PSC, PHR, and SHR data. The multiple regions identified, and their exploration to identify candidate genes, contribute not only to the understanding of the genetic basis of resistance, but also to the development of tools for assisted breeding.
Keywords: Diaporthe/Phomopsis; GWAS; Sclerotinia sclerotiorum; sunflower.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- USDA Oilseeds: World Markets and Trade. [(accessed on 1 August 2022)]; Available online: https://apps.fas.usda.gov/psdonline/circulars/oilseeds.pdf.
-
- Gulya T., Rashid K.Y., Masirevic S.M. Sunflower Diseases. In: Schneiter A.A., editor. Sunflower Technology and Production. American Society of Agronomy; Madison, WI, USA: Crop Science Society of America; Madison, WI, USA: Soil Science Society of America; Madison, WI, USA: 1997. pp. 263–379. Agronomy Monographs.
-
- Gulya T., Harveson R., Mathew F., Block C., Thompson S., Kandel H., Berglund D., Sandbakken J., Kleingartner L., Markell S. Comprehensive disease survey of U.S. sunflower: Disease trends, research priorities and unanticipated impacts. Plant Dis. 2019;103:601–618. doi: 10.1094/PDIS-06-18-0980-FE. - DOI - PubMed
-
- Masirevic S., Gulya T.J. Sclerotinia and Phomopsis—Two devastating sunflower pathogens. Field Crops Res. 1992;30:271–300. doi: 10.1016/0378-4290(92)90004-S. - DOI
-
- Boland G.J., Hall R. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 1994;16:93–108. doi: 10.1080/07060669409500766. - DOI
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

