A Framework for Comparison and Assessment of Synthetic RNA-Seq Data
- PMID: 36553629
- PMCID: PMC9778097
- DOI: 10.3390/genes13122362
A Framework for Comparison and Assessment of Synthetic RNA-Seq Data
Abstract
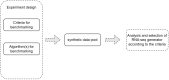
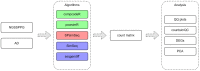
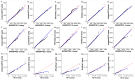
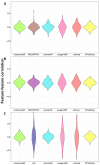
The ever-growing number of methods for the generation of synthetic bulk and single cell RNA-seq data have multiple and diverse applications. They are often aimed at benchmarking bioinformatics algorithms for purposes such as sample classification, differential expression analysis, correlation and network studies and the optimization of data integration and normalization techniques. Here, we propose a general framework to compare synthetically generated RNA-seq data and select a data-generating tool that is suitable for a set of specific study goals. As there are multiple methods for synthetic RNA-seq data generation, researchers can use the proposed framework to make an informed choice of an RNA-seq data simulation algorithm and software that are best suited for their specific scientific questions of interest.
Keywords: RNA-seq; comparative study; differential expression; sample classification; simulated data.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Benchmarking RNA-seq differential expression analysis methods using spike-in and simulation data.PLoS One. 2020 Apr 30;15(4):e0232271. doi: 10.1371/journal.pone.0232271. eCollection 2020. PLoS One. 2020. PMID: 32353015 Free PMC article.
-
Benchmarking differential expression analysis tools for RNA-Seq: normalization-based vs. log-ratio transformation-based methods.BMC Bioinformatics. 2018 Jul 18;19(1):274. doi: 10.1186/s12859-018-2261-8. BMC Bioinformatics. 2018. PMID: 30021534 Free PMC article.
-
SimBA: A methodology and tools for evaluating the performance of RNA-Seq bioinformatic pipelines.BMC Bioinformatics. 2017 Sep 29;18(1):428. doi: 10.1186/s12859-017-1831-5. BMC Bioinformatics. 2017. PMID: 28969586 Free PMC article.
-
Demystifying emerging bulk RNA-Seq applications: the application and utility of bioinformatic methodology.Brief Bioinform. 2021 Nov 5;22(6):bbab259. doi: 10.1093/bib/bbab259. Brief Bioinform. 2021. PMID: 34329375 Review.
-
Evaluation of single-cell classifiers for single-cell RNA sequencing data sets.Brief Bioinform. 2020 Sep 25;21(5):1581-1595. doi: 10.1093/bib/bbz096. Brief Bioinform. 2020. PMID: 31675098 Free PMC article. Review.
Cited by
-
Challenges and best practices in omics benchmarking.Nat Rev Genet. 2024 May;25(5):326-339. doi: 10.1038/s41576-023-00679-6. Epub 2024 Jan 12. Nat Rev Genet. 2024. PMID: 38216661 Review.
-
Special Issue: New Advances in Bioinformatics and Biomedical Engineering Using Machine Learning Techniques, IWBBIO-2022.Genes (Basel). 2023 Aug 1;14(8):1574. doi: 10.3390/genes14081574. Genes (Basel). 2023. PMID: 37628626 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

