Potential Cytoprotective and Regulatory Effects of Ergothioneine on Gene Expression of Proteins Involved in Erythroid Adaptation Mechanisms and Redox Pathways in K562 Cells
- PMID: 36553634
- PMCID: PMC9778224
- DOI: 10.3390/genes13122368
Potential Cytoprotective and Regulatory Effects of Ergothioneine on Gene Expression of Proteins Involved in Erythroid Adaptation Mechanisms and Redox Pathways in K562 Cells
Abstract
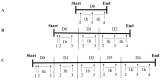
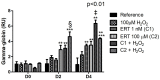
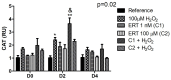
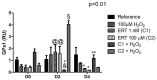
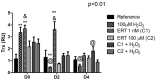
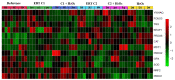
This study aimed to establish the importance of ergothioneine (ERT) in the erythroid adaptation mechanisms by appraising the expression levels of redox-related genes associated with the PI3K/AKT/FoxO3 and Nrf2-ARE pathways using K562 cells induced to erythroid differentiation and H2O2-oxidative stress. Cell viability and gene expression were evaluated. Two concentrations of ERT were assessed, 1 nM (C1) and 100 µM (C2), with and without stress induction (100 µM H2O2). Assessments were made in three periods of the cellular differentiation process (D0, D2, and D4). The C1 treatment promoted the induction of FOXO3 (D0 and 2), PSMB5, and 6 expressions (D4); C1 + H2O2 treatment showed the highest levels of NRF2 transcripts, KEAP1 (D0), YWHAQ (D2 and 4), PSMB5 (D2) and PSMB6 (D4); and C2 + H2O2 (D2) an increase in FOXO3 and MST1 expression, with a decrease of YWHAQ and NRF2 was observed. in C2 + H2O2 (D2) an increase in FOXO3 and MST1, with a decrease in YWHAQ and NRF2 was observed All ERT treatments increased gamma-globin expression. Statistical multivariate analyzes highlighted that the Nrf2-ARE pathway presented a greater contribution in the production of PRDX1, SOD1, CAT, and PSBM5 mRNAs, whereas the PI3K/AKT/FoxO3 pathway was associated with the PRDX2 and TRX transcripts. In conclusion, ERT presented a cytoprotective action through Nrf2 and FoxO3, with the latter seeming to contribute to erythroid proliferation/differentiation.
Keywords: antioxidants; proteasome; redox adaptation; therapeutic agent.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
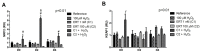



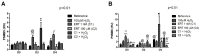
Similar articles
-
Influence of Melatonin Treatment on Cellular Mechanisms of Redox Adaptation in K562 Erythroleukemic Cells.Genes (Basel). 2022 Dec 11;13(12):2337. doi: 10.3390/genes13122337. Genes (Basel). 2022. PMID: 36553603 Free PMC article.
-
Baicalein protects human vitiligo melanocytes from oxidative stress through activation of NF-E2-related factor2 (Nrf2) signaling pathway.Free Radic Biol Med. 2018 Dec;129:492-503. doi: 10.1016/j.freeradbiomed.2018.10.421. Epub 2018 Oct 18. Free Radic Biol Med. 2018. PMID: 30342186
-
Orientin-mediated Nrf2/HO-1 signal alleviates H2O2-induced oxidative damage via induction of JNK and PI3K/AKT activation.Int J Biol Macromol. 2018 Oct 15;118(Pt A):747-755. doi: 10.1016/j.ijbiomac.2018.06.130. Epub 2018 Jun 27. Int J Biol Macromol. 2018. PMID: 29959995
-
Shaping the Nrf2-ARE-related pathways in Alzheimer's and Parkinson's diseases.Ageing Res Rev. 2019 Sep;54:100942. doi: 10.1016/j.arr.2019.100942. Epub 2019 Aug 12. Ageing Res Rev. 2019. PMID: 31415806 Review.
-
Stress-sensing mechanisms and the physiological roles of the Keap1-Nrf2 system during cellular stress.J Biol Chem. 2017 Oct 13;292(41):16817-16824. doi: 10.1074/jbc.R117.800169. Epub 2017 Aug 24. J Biol Chem. 2017. PMID: 28842501 Free PMC article. Review.
Cited by
-
The Biology and Biochemistry of Kynurenic Acid, a Potential Nutraceutical with Multiple Biological Effects.Int J Mol Sci. 2024 Aug 21;25(16):9082. doi: 10.3390/ijms25169082. Int J Mol Sci. 2024. PMID: 39201768 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

