A Whole-Genome Sequencing Study Implicates GRAMD1B in Multiple Sclerosis Susceptibility
- PMID: 36553660
- PMCID: PMC9777893
- DOI: 10.3390/genes13122392
A Whole-Genome Sequencing Study Implicates GRAMD1B in Multiple Sclerosis Susceptibility
Abstract
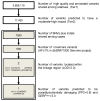
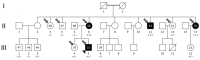
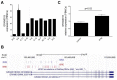
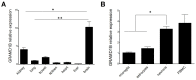
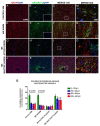
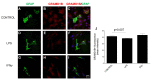
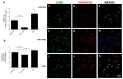
While the role of common genetic variants in multiple sclerosis (MS) has been elucidated in large genome-wide association studies, the contribution of rare variants to the disease remains unclear. Herein, a whole-genome sequencing study in four affected and four healthy relatives of a consanguineous Italian family identified a novel missense c.1801T > C (p.S601P) variant in the GRAMD1B gene that is shared within MS cases and resides under a linkage peak (LOD: 2.194). Sequencing GRAMD1B in 91 familial MS cases revealed two additional rare missense and two splice-site variants, two of which (rs755488531 and rs769527838) were not found in 1000 Italian healthy controls. Functional studies demonstrated that GRAMD1B, a gene with unknown function in the central nervous system (CNS), is expressed by several cell types, including astrocytes, microglia and neurons as well as by peripheral monocytes and macrophages. Notably, GRAMD1B was downregulated in vessel-associated astrocytes of active MS lesions in autopsied brains and by inflammatory stimuli in peripheral monocytes, suggesting a possible role in the modulation of inflammatory response and disease pathophysiology.
Keywords: multiple sclerosis; neurology; rare variants; sequencing.
Conflict of interest statement
Federica Esposito has received honoraria from Serono Symposia International Foundation. A. Dessa Sadovnick has received funding from Biogen and Ronald S. Roadburg Foundation, travel expenses from Biogen and honoraria from Biogen, Novartis, Momenta Pharmaceuticals, and Synofi. Giancarlo Comi has received consulting fees for participating on advisory boards from Novartis, Teva Pharmaceutical Ind. Ltd., Sanofi-Aventis, Merck Serono, Bayer Schering, Actelion and Geneuro and lecture fees from Novartis, Teva Pharmaceutical Industries Ltd., Sanofi-Aventis, Merck Serono, Biogen Dompè, Bayer Schering and Serono Symposia International Foundation. Filippo Martinelli Boneschi has received honoraria for consulting, research grant funding and travel expenses from TEVA Neuroscience, Biogen IDEC, Serono Symposia International Foundation and Genzyme Europe.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

