Identification of Reliable Reference Genes under Different Stresses and in Different Tissues of Toxicodendron succedaneum
- PMID: 36553662
- PMCID: PMC9778191
- DOI: 10.3390/genes13122396
Identification of Reliable Reference Genes under Different Stresses and in Different Tissues of Toxicodendron succedaneum
Abstract
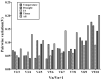
Toxicodendron succedaneum (L.) Kuntze (T. succedaneum) is an economic tree species that produces urushiol and urushi wax, and it is of great value in industry and medicine. However, the stability of reference genes (RGs) has not been systematically reported in T. succedaneum to date. In this study, the expression of 10 candidate RGs was analyzed by RT-qPCR in different tissues (roots, stems, leaves), stress treatments (high/low temperature, drought), and hormone stimulation (jasmonic acid, JA). Then, the stability ranking of 10 candidate genes was evaluated by ∆Ct analysis and three software programs: geNorm, NormFinder, and BestKeeper. Finally, RefFinder was used to comprehensively analyze the expression stability of 10 candidate genes. The comprehensive analysis showed that TsRG05/06, TsRG01/06, and TsRG03/ACT were stable under high/low-temperature stress, drought stress, and JA treatment, respectively. TsRG03 and ACT had stable expression in different tissues. While the TsRG03 and ACT were recommended as the suitable RGs for T. succedaneum in all samples. Meanwhile, UBQ was the least suitable as a reference gene for T. succedaneum. In addition, the results of geNorm showed that the combination of two stable RGs could make the results of gene expression more accurate. These results provide alternative RGs for the study of gene function, correction, and normalization of target gene expression and directed molecular breeding in T. succedaneum.
Keywords: RT-qPCR; Toxicodendron succedaneum; gene expression; reference gene.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Identification and evaluation of reliable reference genes for quantitative real-time PCR analysis in tea plants under differential biotic stresses.Sci Rep. 2020 Feb 12;10(1):2429. doi: 10.1038/s41598-020-59168-z. Sci Rep. 2020. PMID: 32051495 Free PMC article.
-
Selection and validation of appropriate reference genes for RT-qPCR analysis of Nitraria sibirica under various abiotic stresses.BMC Plant Biol. 2022 Dec 17;22(1):592. doi: 10.1186/s12870-022-03988-w. BMC Plant Biol. 2022. PMID: 36526980 Free PMC article.
-
Selection and Validation of Reference Genes for Gene Expression in Bactericera gobica Loginova under Different Insecticide Stresses.Int J Mol Sci. 2024 Feb 19;25(4):2434. doi: 10.3390/ijms25042434. Int J Mol Sci. 2024. PMID: 38397109 Free PMC article.
-
Transcriptome-Based Selection and Validation of Reference Genes for Gene Expression in Goji Fruit Fly (Neoceratitis asiatica Becker) under Developmental Stages and Five Abiotic Stresses.Int J Mol Sci. 2022 Dec 27;24(1):451. doi: 10.3390/ijms24010451. Int J Mol Sci. 2022. PMID: 36613890 Free PMC article.
-
Selection and Optimization of Reference Genes for MicroRNA Expression Normalization by qRT-PCR in Chinese Cedar (Cryptomeria fortunei) under Multiple Stresses.Int J Mol Sci. 2021 Jul 6;22(14):7246. doi: 10.3390/ijms22147246. Int J Mol Sci. 2021. PMID: 34298866 Free PMC article.
Cited by
-
Selection of Reference Genes in Siraitia siamensis and Expression Patterns of Genes Involved in Mogrosides Biosynthesis.Plants (Basel). 2024 Sep 2;13(17):2449. doi: 10.3390/plants13172449. Plants (Basel). 2024. PMID: 39273933 Free PMC article.
References
-
- Ahmed U., Xie Q., Shi X., Zheng B. Development of RGs for Horticultural Plants. Crit. Rev. Plant Sci. 2022;41:190–208. doi: 10.1080/07352689.2022.2084227. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

