Morphological Characteristics and Comparative Chloroplast Genome Analyses between Red and White Flower Phenotypes of Pyracantha fortuneana (Maxim.) Li (Rosaceae), with Implications for Taxonomy and Phylogeny
- PMID: 36553671
- PMCID: PMC9778009
- DOI: 10.3390/genes13122404
Morphological Characteristics and Comparative Chloroplast Genome Analyses between Red and White Flower Phenotypes of Pyracantha fortuneana (Maxim.) Li (Rosaceae), with Implications for Taxonomy and Phylogeny
Abstract
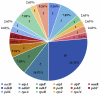
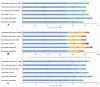
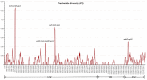
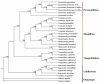
Pyracantha fortuneana (Maxim.) Li (Rosaceae), commonly known as Chinese firethorn, is an evergreen shrub with high nutritional, medicinal, and horticultural importance. This species typically has white flowers, but a rare red flower phenotype has been found in very few wild populations in western Hubei, China, showing great ornamental potential. In this study, the complete chloroplast genome of the red flower phenotype of P. fortuneana was reported for the first time, using high-throughput sequencing technology. The complete chloroplast genome was 160,361 bp in length and showed a typical quadripartite structure with a pair of inverted repeat (IR) regions (26,350 bp) separated by a large single-copy (LSC) region (88,316 bp) and a small single-copy (SSC) region (19,345 bp). A total of 131 functional genes were annotated in this chloroplast genome, including 86 protein-coding genes (PCGs), eight rRNA genes, and 37 tRNA genes. Comparative chloroplast genome analyses revealed that high genome similarity existed not only between red and white flower phenotypes of P. fortuneana, but also among Pyracantha species. No evidence for positive selection was found in any PCG, suggesting the evolutionary conservation of Pyracantha chloroplast genomes. Furthermore, four mutational hotspots (trnG-trnR-atpA, psbZ-trnG-trnfM-rps14, ycf3-trnS-rps4, and ndhF-rpl32) with π > 0.004 were identified as potential molecular markers for Pyracantha species. Phylogenomic analysis strongly supported that the red flower phenotype of P. fortuneana was nested within the common white flower phenotype. Based on both morphological and molecular evidence, we suggest that the red flower phenotype of P. fortuneana could be considered as a new forma. Overall, the availability of these genetic resources will not only offer valuable information for further studies on molecular taxonomy, phylogeny, and population genetics of Pyracantha species but also could be used as potential genetic resources for Chinese firethorn breeding.
Keywords: Pyracantha fortuneana; chloroplast genome; comparative analysis; red flower phenotype; taxonomic investigation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Comparative analysis of the complete chloroplast genomes of thirteen Bougainvillea cultivars from South China with implications for their genome structures and phylogenetic relationships.PLoS One. 2024 Sep 11;19(9):e0310091. doi: 10.1371/journal.pone.0310091. eCollection 2024. PLoS One. 2024. PMID: 39259741 Free PMC article.
-
A systematic comparison of eight new plastome sequences from Ipomoea L.PeerJ. 2019 Mar 11;7:e6563. doi: 10.7717/peerj.6563. eCollection 2019. PeerJ. 2019. PMID: 30881765 Free PMC article.
-
New Insights into Phylogenetic Relationship of Hydrocotyle (Araliaceae) Based on Plastid Genomes.Int J Mol Sci. 2023 Nov 22;24(23):16629. doi: 10.3390/ijms242316629. Int J Mol Sci. 2023. PMID: 38068952 Free PMC article.
-
[Chloroplast genome phylogeny and codon preference of Docynia longiunguis].Sheng Wu Gong Cheng Xue Bao. 2022 Jan 25;38(1):328-342. doi: 10.13345/j.cjb.210298. Sheng Wu Gong Cheng Xue Bao. 2022. PMID: 35142140 Chinese.
-
Dissecting the Biology of Rafflesia Species: Current Progress and Future Directions Made Possible with High-Throughput Sequencing Data.Plant Cell Physiol. 2023 Apr 17;64(4):368-377. doi: 10.1093/pcp/pcad004. Plant Cell Physiol. 2023. PMID: 36611267 Review.
Cited by
-
Advancing ornamental plant breeding through genomic technologies: opportunities, challenges, and future directions.Funct Integr Genomics. 2025 Jul 1;25(1):140. doi: 10.1007/s10142-025-01640-y. Funct Integr Genomics. 2025. PMID: 40590986 Review.
References
-
- Yu D.J., Jiang W.F. Rosaceae. In: Yu D.J., editor. Flora Reipublicae Popularis Sinicae. 2nd ed. Volume 3. Science Press; Beijing, China: 1974. pp. 179–186.
-
- Gu C.Z., Stephen A.S. Rosaceae. In: Wu Z.Y., Raven P.H., editors. Flora of China. 2nd ed. Volume 9. Science Press; Beijing, China: Missouri Botanical Garden Press; St. Louis, MI, USA: 2003. pp. 108–111.
-
- Morgan D.R., Soltis D.E., Robertson K.R. Systematic and evolutionary implications of rbcL sequence variation in Rosaceae. Am. J. Bot. 1994;81:890–903. doi: 10.1002/j.1537-2197.1994.tb15570.x. - DOI
-
- Potter D., Eriksson T., Evans R.C., Oh S., Smedmark J.E.E., Morgan D.R., Kerr M., Robertson K.R., Arsenault M., Dickinson T.A., et al. Phylogeny and classification of Rosaceae. Plant Syst. Evol. 2007;266:5–43. doi: 10.1007/s00606-007-0539-9. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

