Identification of Differentially Expressed miRNAs in Porcine Adipose Tissues and Evaluation of Their Effects on Feed Efficiency
- PMID: 36553673
- PMCID: PMC9778086
- DOI: 10.3390/genes13122406
Identification of Differentially Expressed miRNAs in Porcine Adipose Tissues and Evaluation of Their Effects on Feed Efficiency
Abstract
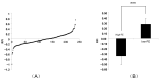
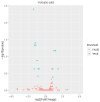
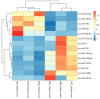
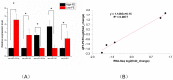
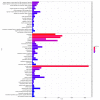
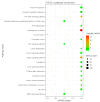
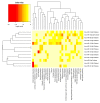
Feed efficiency (FE) is a very important trait affecting the economic benefits of pig breeding enterprises. Adipose tissue can modulate a variety of processes such as feed intake, energy metabolism and systemic physiological processes. However, the mechanism by which microRNAs (miRNAs) in adipose tissues regulate FE remains largely unknown. Therefore, this study aimed to screen potential miRNAs related to FE through miRNA sequencing. The miRNA profiles in porcine adipose tissues were obtained and 14 miRNAs were identified differentially expressed in adipose tissues of pigs with extreme differences in FE, of which 9 were down-regulated and 5 were up-regulated. GO and KEGG analyses indicated that these miRNAs were significantly related to lipid metabolism and these miRNAs modulated FE by regulating lipid metabolism. Subsequently, quantitative reverse transcription-polymerase chain reaction (qRT-PCR) of five randomly selected DEMs was used to verify the reliability of miRNA-seq data. Furthermore, 39 differentially expressed target genes of these DEMs were obtained, and DEMs-target mRNA interaction networks were constructed. In addition, the most significantly down-regulated miRNAs, ssc-miR-122-5p and ssc-miR-192, might be the key miRNAs for FE. Our results reveal the mechanism by which adipose miRNAs regulate feed efficiency in pigs. This study provides a theoretical basis for the further study of swine feed efficiency improvement.
Keywords: adipose tissues; feed efficiency; microRNA; pig; residual feed intake.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Differential expression profile of miRNAs in porcine muscle and adipose tissue during development.Gene. 2017 Jun 30;618:49-56. doi: 10.1016/j.gene.2017.04.013. Epub 2017 Apr 8. Gene. 2017. PMID: 28400270
-
Differences in liver microRNA profiling in pigs with low and high feed efficiency.J Anim Sci Technol. 2022 Mar;64(2):312-329. doi: 10.5187/jast.2022.e4. Epub 2022 Mar 31. J Anim Sci Technol. 2022. PMID: 35530409 Free PMC article.
-
Transcriptome analysis of mRNA and miRNA in skeletal muscle indicates an important network for differential Residual Feed Intake in pigs.Sci Rep. 2015 Jul 7;5:11953. doi: 10.1038/srep11953. Sci Rep. 2015. PMID: 26150313 Free PMC article.
-
Identification of Genes Related to Growth and Lipid Deposition from Transcriptome Profiles of Pig Muscle Tissue.PLoS One. 2015 Oct 27;10(10):e0141138. doi: 10.1371/journal.pone.0141138. eCollection 2015. PLoS One. 2015. PMID: 26505482 Free PMC article.
-
Identification and Characterization of CircRNAs of Two Pig Breeds as a New Biomarker in Metabolism-Related Diseases.Cell Physiol Biochem. 2018;47(6):2458-2470. doi: 10.1159/000491619. Epub 2018 Jul 10. Cell Physiol Biochem. 2018. PMID: 29990990
Cited by
-
Identification of trait-associated microRNA modules in liver transcriptome of pig fed with PUFAs-enriched supplementary diet.J Appl Genet. 2025 May;66(2):389-407. doi: 10.1007/s13353-024-00912-w. Epub 2024 Nov 15. J Appl Genet. 2025. PMID: 39546271 Free PMC article.
References
-
- Hume D.A., Whitelaw C.B.A., Archibald A.L. Foresight project on global food and farming futures the future of animal production: Improving productivity and sustainability. J. Anim. Sci. 2011;149:9–16.
-
- Koch R.L.A.S., Chambers D.K.E.G. Efficiency of feed use in beef cattle. J. Anim. Sci. 1963;22:486–494. doi: 10.2527/jas1963.222486x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

