In Silico Prediction of Hub Genes Involved in Diabetic Kidney and COVID-19 Related Disease by Differential Gene Expression and Interactome Analysis
- PMID: 36553678
- PMCID: PMC9778100
- DOI: 10.3390/genes13122412
In Silico Prediction of Hub Genes Involved in Diabetic Kidney and COVID-19 Related Disease by Differential Gene Expression and Interactome Analysis
Abstract
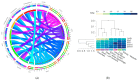
Diabetic kidney disease (DKD) is a frequently chronic kidney pathology derived from diabetes comorbidity. This condition has irreversible damage and its risk factor increases with SARS-CoV-2 infection. The prognostic outcome for diabetic patients with COVID-19 is dismal, even with intensive medical treatment. However, there is still scarce information on critical genes involved in the pathophysiological impact of COVID-19 on DKD. Herein, we characterize differential expression gene (DEG) profiles and determine hub genes undergoing transcriptional reprogramming in both disease conditions. Out of 995 DEGs, we identified 42 shared with COVID-19 pathways. Enrichment analysis elucidated that they are significantly induced with implications for immune and inflammatory responses. By performing a protein-protein interaction (PPI) network and applying topological methods, we determine the following five hub genes: STAT1, IRF7, ISG15, MX1 and OAS1. Then, by network deconvolution, we determine their co-expressed gene modules. Moreover, we validate the conservancy of their upregulation using the Coronascape database (DB). Finally, tissue-specific regulation of the five predictive hub genes indicates that OAS1 and MX1 expression levels are lower in healthy kidney tissue. Altogether, our results suggest that these genes could play an essential role in developing severe outcomes of COVID-19 in DKD patients.
Keywords: COVID-19; diabetic kidney disease; hub genes; in silico analysis; metabolic pathways; potential therapeutic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Diabetic kidney disease-predisposing proinflammatory and profibrotic genes identified by weighted gene co-expression network analysis (WGCNA).J Cell Biochem. 2022 Feb;123(2):481-492. doi: 10.1002/jcb.30195. Epub 2021 Dec 14. J Cell Biochem. 2022. PMID: 34908186
-
Identification of Novel Key Molecular Signatures in the Pathogenesis of Experimental Diabetic Kidney Disease.Front Endocrinol (Lausanne). 2022 Mar 30;13:843721. doi: 10.3389/fendo.2022.843721. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35432190 Free PMC article.
-
Bioinformatics prediction and experimental verification of key biomarkers for diabetic kidney disease based on transcriptome sequencing in mice.PeerJ. 2022 Sep 20;10:e13932. doi: 10.7717/peerj.13932. eCollection 2022. PeerJ. 2022. PMID: 36157062 Free PMC article.
-
5-Hydroxymethylcytosine profiles in plasma cell-free DNA reflect molecular characteristics of diabetic kidney disease.Front Endocrinol (Lausanne). 2022 Jul 29;13:910907. doi: 10.3389/fendo.2022.910907. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35966076 Free PMC article.
-
An update on the interaction between COVID-19, vaccines, and diabetic kidney disease.Front Immunol. 2022 Oct 20;13:999534. doi: 10.3389/fimmu.2022.999534. eCollection 2022. Front Immunol. 2022. PMID: 36341356 Free PMC article. Review.
Cited by
-
Circulating Factors as Potential Biomarkers of Cardiovascular Damage Progression Associated with Type 2 Diabetes.Proteomes. 2024 Oct 11;12(4):29. doi: 10.3390/proteomes12040029. Proteomes. 2024. PMID: 39449501 Free PMC article.
-
Predicting the hub interactome of COVID-19 and oral squamous cell carcinoma: uncovering ALDH-mediated Wnt/β-catenin pathway activation via salivary inflammatory proteins.Sci Rep. 2025 Feb 3;15(1):4068. doi: 10.1038/s41598-025-88819-2. Sci Rep. 2025. PMID: 39901050 Free PMC article.
-
Interferon-Stimulated Genes: Novel Targets in Renal Pathogenesis.Kidney Dis (Basel). 2025 May 2;11(1):390-401. doi: 10.1159/000546141. eCollection 2025 Jan-Dec. Kidney Dis (Basel). 2025. PMID: 40519215 Free PMC article. Review.
-
Integration of Omics Data and Network Models to Unveil Negative Aspects of SARS-CoV-2, from Pathogenic Mechanisms to Drug Repurposing.Biology (Basel). 2023 Aug 31;12(9):1196. doi: 10.3390/biology12091196. Biology (Basel). 2023. PMID: 37759595 Free PMC article. Review.
-
Gene regulatory networks in disease and ageing.Nat Rev Nephrol. 2024 Sep;20(9):616-633. doi: 10.1038/s41581-024-00849-7. Epub 2024 Jun 12. Nat Rev Nephrol. 2024. PMID: 38867109 Review.
References
-
- Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

