Proteomic Profiling of Major Peanut Allergens and Their Post-Translational Modifications Affected by Roasting
- PMID: 36553735
- PMCID: PMC9778155
- DOI: 10.3390/foods11243993
Proteomic Profiling of Major Peanut Allergens and Their Post-Translational Modifications Affected by Roasting
Abstract
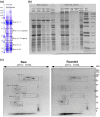
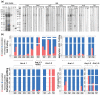
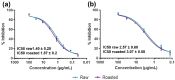
Post-translational modifications (PTMs) are covalent changes occurring on amino acid side chains of proteins and yet are neglected structural and functional aspects of protein architecture. The objective was to detect differences in PTM profiles that take place after roasting using open PTM search. We conducted a bottom-up proteomic study to investigate the impact of peanut roasting on readily soluble allergens and their PTM profiles. Proteomic PTM profiling of certain modifications was confirmed by Western blotting with a series of PTM-specific antibodies. In addition to inducing protein aggregation and denaturation, roasting may facilitate change in their PTM pattern and relative profiling. We have shown that Ara h 1 is the most modified major allergen in both samples in terms of modification versatility and extent. The most frequent PTM was methionine oxidation, especially in roasted samples. PTMs uniquely found in roasted samples were hydroxylation (Trp), formylation (Arg/Lys), and oxidation or hydroxylation (Asn). Raw and roasted peanut extracts did not differ in the binding of IgE from the serum of peanut-sensitised individuals done by ELISA. This study provides a better understanding of how roasting impacts the PTM profile of major peanut allergens and provides a good foundation for further exploration of PTMs.
Keywords: PTM profiling; allergy; high resolution mass spectrometry (HRMS); peanut allergen profiling; post-translational modifications; roasting; shotgun proteomics; western blot.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Smiljanić K., Prodić I., Apostolović D., Cvetković A., Veljović D., Mutić J., van Hage M., Burazer L., Veličković T.C. In-depth quantitative profiling of post-translational modifications of Timothy grass pollen allergome in relation to environmental oxidative stress. Environ. Int. 2019;126:644–658. doi: 10.1016/j.envint.2019.03.001. - DOI - PubMed
-
- Smiljanić K., Mihailović J., Prodić I., Đukić T., Vasović T., Jovanović V., Veličković T.C. Trypsin as a Proteomic Probe for Assessment of Food Protein Digestibility in Relation to Chemical and Post-Translational Modifications. In: Radosavljevic J., editor. A Closer Look at Proteolysis. 1st ed. Nova Science Publishers, Inc.; New York, NY, USA: 2020. pp. 157–184.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

