Transcriptome Analysis and Identification of Chemosensory Genes in Baryscapus dioryctriae (Hymenoptera: Eulophidae)
- PMID: 36555008
- PMCID: PMC9780838
- DOI: 10.3390/insects13121098
Transcriptome Analysis and Identification of Chemosensory Genes in Baryscapus dioryctriae (Hymenoptera: Eulophidae)
Abstract
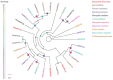
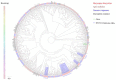
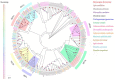
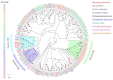
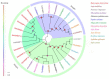
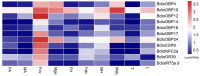
Baryscapus dioryctriae is a pupal endoparasitoid of many Pyralidae pests and has been used as a biocontrol agent against insect pests that heavily damage the cone and seed of the Korean pine. The olfactory system of wasps plays an essential role in sensing the chemical signals during their foraging, mating, host location, etc., and the chemosensory genes are involved in detecting and transducing these signals. Many chemosensory genes have been identified from the antennae of Hymenoptera; however, there are few reports on the chemosensory genes of Eulophidae wasps. In this study, the transcriptome databases based on ten different tissues of B. dioryctriae were first constructed, and 274 putative chemosensory genes, consisting of 27 OBPs, 9 CSPs, 3 NPC2s, 155 ORs, 49 GRs, 23 IRs and 8 SNMPs genes, were identified based on the transcriptomes and manual annotation. Phylogenetic trees of the chemosensory genes were constructed to investigate the orthologs between B. dioryctriae and other insect species. Additionally, twenty-eight chemosensory genes showed female antennae- and ovipositor-biased expression, which was validated by RT-qPCR. These findings not only built a molecular basis for further research on the processes of chemosensory perception in B. dioryctriae, but also enriched the identification of chemosensory genes from various tissues of Eulophidae wasps.
Keywords: Baryscapus dioryctriae; Eulophidae; chemosensory genes; transcriptome; wasp.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
An expanded odorant-binding protein mediates host cue detection in the parasitic wasp Baryscapus dioryctriae basis of the chromosome-level genome assembly analysis.BMC Biol. 2024 Sep 11;22(1):196. doi: 10.1186/s12915-024-01998-8. BMC Biol. 2024. PMID: 39256805 Free PMC article.
-
Morphology and ultrastructure of antennal sensilla of the parasitic wasp Baryscapus dioryctriae (Hymenoptera: Eulophidae).Microsc Res Tech. 2023 Jan;86(1):12-27. doi: 10.1002/jemt.24253. Epub 2022 Nov 1. Microsc Res Tech. 2023. PMID: 36318186
-
Identification and analysis of olfactory genes in Dioryctria abietella based on the antennal transcriptome.Comp Biochem Physiol Part D Genomics Proteomics. 2021 Jun;38:100814. doi: 10.1016/j.cbd.2021.100814. Epub 2021 Feb 15. Comp Biochem Physiol Part D Genomics Proteomics. 2021. PMID: 33706113
-
Identification and sex-biased profiles of candidate olfactory genes in the antennal transcriptome of the parasitoid wasp Cotesia vestalis.Comp Biochem Physiol Part D Genomics Proteomics. 2020 Jun;34:100657. doi: 10.1016/j.cbd.2020.100657. Epub 2020 Jan 29. Comp Biochem Physiol Part D Genomics Proteomics. 2020. PMID: 32032903
-
Functional Characterization of the Niemann-Pick C2 Protein BdioNPC2b in the Parasitic Wasp Baryscapus dioryctriae (Chalcidodea: Eulophidae).J Agric Food Chem. 2024 Apr 10;72(14):7735-7748. doi: 10.1021/acs.jafc.3c09095. Epub 2024 Mar 28. J Agric Food Chem. 2024. PMID: 38546111
Cited by
-
Identification and expression profiles of olfactory-related genes in the antennal transcriptome of Graphosoma rubrolineatum (Hemiptera: Pentatomidae).PLoS One. 2024 Aug 6;19(8):e0306986. doi: 10.1371/journal.pone.0306986. eCollection 2024. PLoS One. 2024. PMID: 39106289 Free PMC article.
-
Identification of candidate genes associated with host-seeking behavior in the parasitoid wasp Diachasmimorpha longicaudata.BMC Genomics. 2024 Feb 6;25(1):147. doi: 10.1186/s12864-024-10034-6. BMC Genomics. 2024. PMID: 38321385 Free PMC article.
-
The Olfactory System of Dolichogenidea gelechiidivoris (Marsh) (Hymenoptera: Braconidae), a Natural Enemy of Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae).Int J Mol Sci. 2025 Jul 29;26(15):7312. doi: 10.3390/ijms26157312. Int J Mol Sci. 2025. PMID: 40806444 Free PMC article.
-
Transcriptome analysis and identification of genes related to environmental adaptation of Grylloprimevala jilina Zhou & Ren 2023.Ecol Evol. 2023 Nov 20;13(11):e10717. doi: 10.1002/ece3.10717. eCollection 2023 Nov. Ecol Evol. 2023. PMID: 38020696 Free PMC article.
-
An expanded odorant-binding protein mediates host cue detection in the parasitic wasp Baryscapus dioryctriae basis of the chromosome-level genome assembly analysis.BMC Biol. 2024 Sep 11;22(1):196. doi: 10.1186/s12915-024-01998-8. BMC Biol. 2024. PMID: 39256805 Free PMC article.
References
-
- Ali A.N., Wright M.G. Response of Trichogramma papilionis to semiochemicals induced by host oviposition on plants. Biol. Control. 2021;154:104510. doi: 10.1016/j.biocontrol.2020.104510. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

