Transcriptomic Analysis Insight into the Immune Modulation during the Interaction of Ophiocordyceps sinensis and Hepialus xiaojinensis
- PMID: 36555029
- PMCID: PMC9788539
- DOI: 10.3390/insects13121119
Transcriptomic Analysis Insight into the Immune Modulation during the Interaction of Ophiocordyceps sinensis and Hepialus xiaojinensis
Abstract
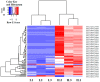
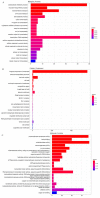
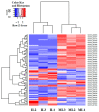
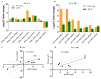
Ophiocordyceps sinensis (Berk.) is an entomopathogenic fungus that can infect the larva of the ghost moth, Hepialus xiaojinensis, causing mummification after more than one year. This prolonged infection provides a valuable model for studying the immunological interplay between an insect host and a pathogenic fungus. A comparative transcriptome analysis of pre-infection (L) and one-year post-infection (IL) larvae was performed to investigate the immune response in the host. Here, a total of 59,668 unigenes were obtained using Illumina Sequencing in IL and L. Among the 345 identified immune-related genes, 83 out of 86 immune-related differentially expressed genes (DEGs) had a much higher expression in IL than in L. Furthermore, the immune-related DEGs were classified as pathogen recognition receptors (PRRs), signal modulators or transductors, and immune effector molecules. Serpins and protease inhibitors were found to be upregulated in the late phase of infection, suppressing the host’s immune response. Based on the above analysis, the expression levels of most immune-related genes would return to the baseline with the immune response being repressed in the late phase of infection, leading to the fungal immunological tolerance after prolonged infection. Meanwhile, the transcriptomes of IL and the mummified larva (ML) were compared to explore O. sinensis invasion. A total of 1408 novel genes were identified, with 162 of them annotated with putative functions. The gene families likely implicated in O. sinensis pathogenicity have been identified, primarily including serine carboxypeptidase, peroxidase, metalloprotease peptidase, aminopeptidases, cytochrome P450, and oxidoreductase. Furthermore, quantitative real-time PCR (qPCR) was used to assess the expression levels of some critical genes that were involved in immune response and fungal pathogenicity. The results showed that their expression levels were consistent with the transcriptomes. Taken together, our findings offered a comprehensive and precise transcriptome study to understand the immune defense in H. xiaojinensis and O. sinensis invasion, which would accelerate the large-scale artificial cultivation of this medicinal fungus.
Keywords: Hepialus xiaojinensis; Ophiocordyceps sinensis; RNA-seqs; immunological interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









References
-
- Wei Y., Zhang L., Wang J., Wang W., Niyati N., Guo Y., Wang X. Chinese caterpillar fungus (Ophiocordyceps sinensis) in China: Current distribution, trading, and futures under climate change and overexploitation. Sci. Total Environ. 2021;755:142548. doi: 10.1016/j.scitotenv.2020.142548. - DOI - PMC - PubMed
-
- Zhang Y.J., Li E., Wang C.S., Li Y.L., Liu X.Z. Ophiocordyceps sinensis, the flagship fungus of China: Terminology, life strategy and ecology. Mycology. 2012;3:2–10.
-
- Guo J.L., Liu X.Y., Kanari K. Economic aspects of harvesting and trading the Chinese Caterpillar Fungus Ophiocordyceps sinensis and Southern Schisandra Schisandra sphenanthera in China’s Upper Yangtze Ecoregion. Traffic Bull. 2012;24:15–24.
Grants and funding
LinkOut - more resources
Full Text Sources

