A Large Intergenic Spacer Leads to the Increase in Genome Size and Sequential Gene Movement around IR/SC Boundaries in the Chloroplast Genome of Adiantum malesianum (Pteridaceae)
- PMID: 36555263
- PMCID: PMC9778900
- DOI: 10.3390/ijms232415616
A Large Intergenic Spacer Leads to the Increase in Genome Size and Sequential Gene Movement around IR/SC Boundaries in the Chloroplast Genome of Adiantum malesianum (Pteridaceae)
Abstract
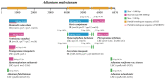
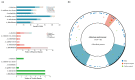
Expansion and contraction (ebb and flow events) of inverted repeat (IR) boundaries occur and are generally considered to be major factors affecting chloroplast (cp) genome size changes. Nonetheless, the Adiantum malesianum cp genome does not seem to follow this pattern. We sequenced, assembled and corrected the A. flabellulatum and A. malesianum cp genomes using the Illumina NovaSeq6000 platform, and we performed a comparative genome analysis of six Adiantum species. The results revealed differences in the IR/SC boundaries of A. malesianum caused by a 6876 bp long rpoB-trnD-GUC intergenic spacer (IGS) in the LSC. This IGS may create topological tension towards the LSC/IRb boundary in the cp genome, resulting in a sequential movement of the LSC genes. Consequently, this leads to changes of the IR/SC boundaries and may even destroy the integrity of trnT-UGU, which is located in IRs. This study provides evidence showing that it is the large rpoB-trnD-GUC IGS that leads to A. malesianum cp genome size change, rather than ebb and flow events. Then, the study provides a model to explain how the rpoB-trnD-GUC IGS in LSC affects A. malesianum IR/SC boundaries. Moreover, this study also provides useful data for dissecting the evolution of cp genomes of Adiantum. In future research, we can expand the sample to Pteridaceae to test whether this phenomenon is universal in Pteridaceae.
Keywords: Adiantum malesianum; chloroplast conformations; genome comparison; inverted repeat; topological tension.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Dobrogojski J., Adamiec M., Luciński R. The chloroplast genome: A review. Acta Physiol. Plant. 2020;42:98. doi: 10.1007/s11738-020-03089-x. - DOI
-
- Bock R. Structure, Function, and Inheritance of Plastid Genomes. Springer; Berlin, Heidelberg: 2007. pp. 29–63.
-
- Menezes A., Resende-Moreira L.C., Buzatti R., Nazareno A.G., Carlsen M., Lobo F.P., Kalapothakis E., Lovato M.B. Chloroplast genomes of Byrsonima species (Malpighiaceae): Comparative analysis and screening of high divergence sequences. Sci. Rep. 2018;8:2210. doi: 10.1038/s41598-018-20189-4. - DOI - PMC - PubMed
MeSH terms
Grants and funding
- 31872670 and 32071781/the National Natural Science Foundation of China
- 2021A1515010911/Guangdong Basic and Applied Basic Research Foundation
- 202206010107/Science and Technology Projects in Guangzhou
- JCYJ20190813172001780 and JCYJ20210324141000001/Project of Department of Science and Technology of Shenzhen City, Guangdong, China
LinkOut - more resources
Full Text Sources
Miscellaneous

