Cost-Effective Mechanical Aggregation of Cardiac Progenitors and Encapsulation in Matrigel Support Self-Organization in a Dynamic Culture Environment
- PMID: 36555427
- PMCID: PMC9779514
- DOI: 10.3390/ijms232415785
Cost-Effective Mechanical Aggregation of Cardiac Progenitors and Encapsulation in Matrigel Support Self-Organization in a Dynamic Culture Environment
Abstract
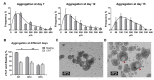
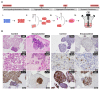
Human iPSC-derived self-organized cardiac tissues can be valuable for the development of platforms for disease modeling and drug screening, enhancing test accuracy and reducing pharmaceutical industry financial burden. However, current differentiation systems still rely on static culture conditions and specialized commercial microwells for aggregation, which hinders the full potential of hiPSC-derived cardiac tissues. Herein, we integrate cost-effective and reproducible manual aggregation of hiPSC-derived cardiac progenitors with Matrigel encapsulation and a dynamic culture to support hiPSC cardiac differentiation and self-organization. Manual aggregation at day 7 of cardiac differentiation resulted in 97% of beating aggregates with 78% of cTnT-positive cells. Matrigel encapsulation conjugated with a dynamic culture promoted cell migration and the creation of organized structures, with observed cell polarization and the creation of lumens. In addition, encapsulation increased buoyancy and decreased coalescence of the hiPSC-derived cardiac aggregates. Moreover, VEGF supplementation increased over two-fold the percentage of CD31-positive cells resulting in the emergence of microvessel-like structures. Thus, this study shows that the explored culture parameters support the self-organization of hiPSC-derived cardiac microtissues containing multiple cardiac cell types. Additional stimuli (e.g., BMP) in long-term scalable and fully automatized cultures can further potentiate highly structured and mature hiPSC-derived cardiac models, contributing to the development of reliable platforms for high-throughput drug screening and disease modeling.
Keywords: Matrigel encapsulation; WNT signaling modulation; cardiac differentiation; dynamic culture; manual aggregation; self-organization.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Matrigel Mattress: A Method for the Generation of Single Contracting Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes.Circ Res. 2015 Dec 4;117(12):995-1000. doi: 10.1161/CIRCRESAHA.115.307580. Epub 2015 Oct 1. Circ Res. 2015. PMID: 26429802 Free PMC article.
-
Thyroid and Glucocorticoid Hormones Promote Functional T-Tubule Development in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes.Circ Res. 2017 Dec 8;121(12):1323-1330. doi: 10.1161/CIRCRESAHA.117.311920. Epub 2017 Oct 2. Circ Res. 2017. PMID: 28974554 Free PMC article.
-
Biophysical study of human induced Pluripotent Stem Cell-Derived cardiomyocyte structural maturation during long-term culture.Biochem Biophys Res Commun. 2018 May 15;499(3):611-617. doi: 10.1016/j.bbrc.2018.03.198. Epub 2018 Mar 30. Biochem Biophys Res Commun. 2018. PMID: 29601816
-
Modeling Cardiovascular Diseases with hiPSC-Derived Cardiomyocytes in 2D and 3D Cultures.Int J Mol Sci. 2020 May 11;21(9):3404. doi: 10.3390/ijms21093404. Int J Mol Sci. 2020. PMID: 32403456 Free PMC article. Review.
-
The effects of xeno-free cryopreservation on the contractile properties of human iPSC derived cardiomyocytes.J Mol Cell Cardiol. 2022 Jul;168:107-114. doi: 10.1016/j.yjmcc.2022.04.010. Epub 2022 Apr 21. J Mol Cell Cardiol. 2022. PMID: 35461823 Review.
Cited by
-
Appraisal of CRISPR Technology as an Innovative Screening to Therapeutic Toolkit for Genetic Disorders.Mol Biotechnol. 2025 Feb 2. doi: 10.1007/s12033-025-01374-z. Online ahead of print. Mol Biotechnol. 2025. PMID: 39894889 Review.
-
Induced Pluripotent Stem Cell-Derived Organoids: Their Implication in COVID-19 Modeling.Int J Mol Sci. 2023 Feb 9;24(4):3459. doi: 10.3390/ijms24043459. Int J Mol Sci. 2023. PMID: 36834870 Free PMC article. Review.
References
-
- Waring M.J., Arrowsmith J., Leach A.R., Leeson P.D., Mandrell S., Owen R.M., Pairaudeau G., Pennie W.D., Pickett S.D., Wang J., et al. An Analysis of the Attrition of Drug Candidates from Four Major Pharmaceutical Companies. Nat. Rev. Drug Discov. 2015;14:475–486. doi: 10.1038/nrd4609. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

